A hand-off of DNA between archaeal polymerases allows high-fidelity replication to resume at a discrete intermediate three bases past 8-oxoguanine
- PMID: 32997110
- PMCID: PMC7641752
- DOI: 10.1093/nar/gkaa803
A hand-off of DNA between archaeal polymerases allows high-fidelity replication to resume at a discrete intermediate three bases past 8-oxoguanine
Abstract
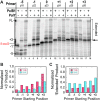
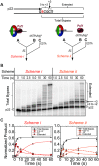
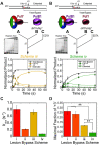
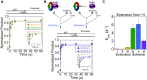
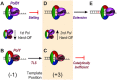
During DNA replication, the presence of 8-oxoguanine (8-oxoG) lesions in the template strand cause the high-fidelity (HiFi) DNA polymerase (Pol) to stall. An early response to 8-oxoG lesions involves 'on-the-fly' translesion synthesis (TLS), in which a specialized TLS Pol is recruited and replaces the stalled HiFi Pol for lesion bypass. The length of TLS must be long enough for effective bypass, but it must also be regulated to minimize replication errors by the TLS Pol. The exact position where the TLS Pol ends and the HiFi Pol resumes (i.e. the length of the TLS patch) has not been described. We use steady-state and pre-steady-state kinetic assays to characterize lesion bypass intermediates formed by different archaeal polymerase holoenzyme complexes that include PCNA123 and RFC. After bypass of 8-oxoG by TLS PolY, products accumulate at the template position three base pairs beyond the lesion. PolY is catalytically poor for subsequent extension from this +3 position beyond 8-oxoG, but this inefficiency is overcome by rapid extension of HiFi PolB1. The reciprocation of Pol activities at this intermediate indicates a defined position where TLS Pol extension is limited and where the DNA substrate is handed back to the HiFi Pol after bypass of 8-oxoG.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Kunkel T.A. DNA replication fidelity. J. Biol. Chem. 2004; 279:16895–16898. - PubMed
-
- Mouron S., Rodriguez-Acebes S., Martinez-Jimenez M.I., Garcia-Gomez S., Chocron S., Blanco L., Mendez J.. Repriming of DNA synthesis at stalled replication forks by human PrimPol. Nat. Struct. Mol. Biol. 2013; 20:1383–1389. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

