A suite of 19F based relaxation dispersion experiments to assess biomolecular motions
- PMID: 32997265
- PMCID: PMC7701166
- DOI: 10.1007/s10858-020-00348-4
A suite of 19F based relaxation dispersion experiments to assess biomolecular motions
Erratum in
-
Correction to: A suite of 19F based relaxation dispersion experiments to assess biomolecular motions.J Biomol NMR. 2020 Dec;74(12):767-768. doi: 10.1007/s10858-020-00352-8. J Biomol NMR. 2020. PMID: 33237374 Free PMC article.
Abstract
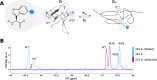
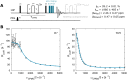
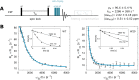
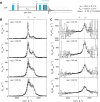
Proteins and nucleic acids are highly dynamic bio-molecules that can populate a variety of conformational states. NMR relaxation dispersion (RD) methods are uniquely suited to quantify the associated kinetic and thermodynamic parameters. Here, we present a consistent suite of 19F-based CPMG, on-resonance R1ρ and off-resonance R1ρ RD experiments. We validate these experiments by studying the unfolding transition of a 7.5 kDa cold shock protein. Furthermore we show that the 19F RD experiments are applicable to very large molecular machines by quantifying dynamics in the 360 kDa half-proteasome. Our approach significantly extends the timescale of chemical exchange that can be studied with 19F RD, adds robustness to the extraction of exchange parameters and can determine the absolute chemical shifts of excited states. Importantly, due to the simplicity of 19F NMR spectra, it is possible to record complete datasets within hours on samples that are of very low costs. This makes the presented experiments ideally suited to complement static structural information from cryo-EM and X-ray crystallography with insights into functionally relevant motions.
Keywords: Fluorine; Large complexes; Protein folding; Relaxation dispersion; Structural dynamics..
Figures


Similar articles
-
Role of entropy in protein thermostability: folding kinetics of a hyperthermophilic cold shock protein at high temperatures using 19F NMR.Biochemistry. 2002 Oct 1;41(39):11670-80. doi: 10.1021/bi026293l. Biochemistry. 2002. PMID: 12269809
-
Resolving biomolecular motion and interactions by R2 and R1ρ relaxation dispersion NMR.Methods. 2018 Sep 15;148:28-38. doi: 10.1016/j.ymeth.2018.04.026. Epub 2018 Apr 26. Methods. 2018. PMID: 29704666 Review.
-
Overview of Relaxation Dispersion NMR Spectroscopy to Study Protein Dynamics and Protein-Ligand Interactions.Curr Protoc Protein Sci. 2018 Apr;92(1):e57. doi: 10.1002/cpps.57. Curr Protoc Protein Sci. 2018. PMID: 30040207
-
Probing invisible, low-populated States of protein molecules by relaxation dispersion NMR spectroscopy: an application to protein folding.Acc Chem Res. 2008 Mar;41(3):442-51. doi: 10.1021/ar700189y. Epub 2008 Feb 15. Acc Chem Res. 2008. PMID: 18275162 Review.
-
NMR Relaxation Dispersion Methods for the Structural and Dynamic Analysis of Quickly Interconverting, Low-Populated Conformational Substates.Methods Mol Biol. 2022;2376:187-203. doi: 10.1007/978-1-0716-1716-8_11. Methods Mol Biol. 2022. PMID: 34845611
Cited by
-
19F NMR Untersuchung des Konformationsaustauschs mehrerer Zustände im synthetischen Neomycin-bindenden Riboschalter.Angew Chem Weinheim Bergstr Ger. 2023 Jun 5;135(23):e202218064. doi: 10.1002/ange.202218064. Epub 2023 Apr 28. Angew Chem Weinheim Bergstr Ger. 2023. PMID: 38516132 Free PMC article.
-
High-Efficiency Trifluoromethyl-Methionine Incorporation into Cyclophilin A by Cell-Free Synthesis for 19F NMR Studies.Angew Chem Int Ed Engl. 2025 Feb 10;64(7):e202419709. doi: 10.1002/anie.202419709. Epub 2024 Dec 4. Angew Chem Int Ed Engl. 2025. PMID: 39571097
-
TRPM8 Protein Dynamics Correlates with Ligand Structure and Cellular Function.J Am Chem Soc. 2025 Jun 4;147(22):18460-18474. doi: 10.1021/jacs.4c09435. Epub 2025 May 27. J Am Chem Soc. 2025. PMID: 40421580 Free PMC article.
-
The ribosome stabilizes partially folded intermediates of a nascent multi-domain protein.Nat Chem. 2022 Oct;14(10):1165-1173. doi: 10.1038/s41557-022-01004-0. Epub 2022 Aug 4. Nat Chem. 2022. PMID: 35927328 Free PMC article.
-
19F NMR viewed through two different lenses: ligand-observed and protein-observed 19F NMR applications for fragment-based drug discovery.RSC Chem Biol. 2021 Jul 12;2(5):1312-1330. doi: 10.1039/d1cb00085c. eCollection 2021 Oct 7. RSC Chem Biol. 2021. PMID: 34704040 Free PMC article. Review.
References
-
- Abraham RJ, Jones AD, Warne MA, et al. Conformational analysis. Part 27. NMR, solvation and theoretical investigation of conformational isomerism in fluoro- and 1,1-difluoro-acetone. J Chem Soc Perkin Trans. 1996;2:533–539. doi: 10.1039/P29960000533. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

