The STRIPAK signaling complex regulates dephosphorylation of GUL1, an RNA-binding protein that shuttles on endosomes
- PMID: 32997654
- PMCID: PMC7550108
- DOI: 10.1371/journal.pgen.1008819
The STRIPAK signaling complex regulates dephosphorylation of GUL1, an RNA-binding protein that shuttles on endosomes
Abstract
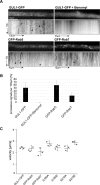
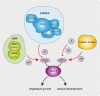
The striatin-interacting phosphatase and kinase (STRIPAK) multi-subunit signaling complex is highly conserved within eukaryotes. In fungi, STRIPAK controls multicellular development, morphogenesis, pathogenicity, and cell-cell recognition, while in humans, certain diseases are related to this signaling complex. To date, phosphorylation and dephosphorylation targets of STRIPAK are still widely unknown in microbial as well as animal systems. Here, we provide an extended global proteome and phosphoproteome study using the wild type as well as STRIPAK single and double deletion mutants (Δpro11, Δpro11Δpro22, Δpp2Ac1Δpro22) from the filamentous fungus Sordaria macrospora. Notably, in the deletion mutants, we identified the differential phosphorylation of 129 proteins, of which 70 phosphorylation sites were previously unknown. Included in the list of STRIPAK targets are eight proteins with RNA recognition motifs (RRMs) including GUL1. Knockout mutants and complemented transformants clearly show that GUL1 affects hyphal growth and sexual development. To assess the role of GUL1 phosphorylation on fungal development, we constructed phospho-mimetic and -deficient mutants of GUL1 residues. While S180 was dephosphorylated in a STRIPAK-dependent manner, S216, and S1343 served as non-regulated phosphorylation sites. While the S1343 mutants were indistinguishable from wild type, phospho-deficiency of S180 and S216 resulted in a drastic reduction in hyphal growth, and phospho-deficiency of S216 also affects sexual fertility. These results thus suggest that differential phosphorylation of GUL1 regulates developmental processes such as fruiting body maturation and hyphal morphogenesis. Moreover, genetic interaction studies provide strong evidence that GUL1 is not an integral subunit of STRIPAK. Finally, fluorescence microscopy revealed that GUL1 co-localizes with endosomal marker proteins and shuttles on endosomes. Here, we provide a new mechanistic model that explains how STRIPAK-dependent and -independent phosphorylation of GUL1 regulates sexual development and asexual growth.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
-
- Rogers S, McCloy R, Watkins DN, Burgess A. Mechanisms regulating phosphatase specificity and the removal of individual phosphorylation sites during mitotic exit. Bioessays. 2016;38 Suppl 1:S24–32. - PubMed
-
- Goudreault M, D'Ambrosio LM, Kean MJ, Mullin MJ, Larsen BG, Sanchez A, et al. A PP2A phosphatase high density interaction network identifies a novel striatin-interacting phosphatase and kinase complex linked to the cerebral cavernous malformation 3 (CCM3) protein. Mol Cell Proteomics. 2009;8(1):157–71. 10.1074/mcp.M800266-MCP200 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

