Effects of germline and somatic events in candidate BRCA-like genes on breast-tumor signatures
- PMID: 32997669
- PMCID: PMC7526916
- DOI: 10.1371/journal.pone.0239197
Effects of germline and somatic events in candidate BRCA-like genes on breast-tumor signatures
Abstract
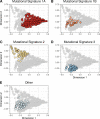
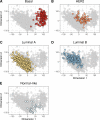
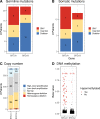
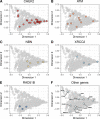
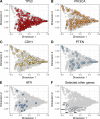
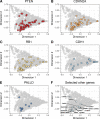
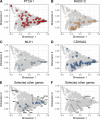
Mutations in BRCA1 and BRCA2 cause deficiencies in homologous recombination repair (HR), resulting in repair of DNA double-strand breaks by the alternative non-homologous end-joining pathway, which is more error prone. HR deficiency of breast tumors is important because it is associated with better responses to platinum salt therapies and PARP inhibitors. Among other consequences of HR deficiency are characteristic somatic-mutation signatures and gene-expression patterns. The term "BRCA-like" (or "BRCAness") describes tumors that harbor an HR defect but have no detectable germline mutation in BRCA1 or BRCA2. A better understanding of the genes and molecular events associated with tumors being BRCA-like could provide mechanistic insights and guide development of targeted treatments. Using data from The Cancer Genome Atlas (TCGA) for 1101 breast-cancer patients, we identified individuals with a germline mutation, somatic mutation, homozygous deletion, and/or hypermethylation event in BRCA1, BRCA2, and 59 other cancer-predisposition genes. Based on the assumption that BRCA-like events would have similar downstream effects on tumor biology as BRCA1/BRCA2 germline mutations, we quantified these effects based on somatic-mutation signatures and gene-expression profiles. We reduced the dimensionality of the somatic-mutation signatures and expression data and used a statistical resampling approach to quantify similarities among patients who had a BRCA1/BRCA2 germline mutation, another type of aberration in BRCA1 or BRCA2, or any type of aberration in one of the other genes. Somatic-mutation signatures of tumors having a non-germline aberration in BRCA1/BRCA2 (n = 80) were generally similar to each other and to tumors from BRCA1/BRCA2 germline carriers (n = 44). Additionally, somatic-mutation signatures of tumors with germline or somatic events in ATR (n = 16) and BARD1 (n = 8) showed high similarity to tumors from BRCA1/BRCA2 carriers. Other genes (CDKN2A, CTNNA1, PALB2, PALLD, PRSS1, SDHC) also showed high similarity but only for a small number of events or for a single event type. Tumors with germline mutations or hypermethylation of BRCA1 had relatively similar gene-expression profiles and overlapped considerably with the Basal-like subtype; but the transcriptional effects of the other events lacked consistency. Our findings confirm previously known relationships between molecular signatures and germline or somatic events in BRCA1/BRCA2. Our methodology represents an objective way to identify genes that have similar downstream effects on molecular signatures when mutated, deleted, or hypermethylated.
Conflict of interest statement
TW consults for Color Genomics. Otherwise, the authors declare that they have no competing interests. This does not alter our adherence to PLOS ONE policies on sharing data and materials.
Figures
References
-
- Malone KE, Daling JR, Doody DR, Hsu L, Bernstein L, Coates RJ, et al. Prevalence and predictors of BRCA1 and BRCA2 mutations in a population-based study of breast cancer in white and black American women ages 35 to 64 years. Cancer Res. 2006. August;66(16):8297–308. 10.1158/0008-5472.CAN-06-0503 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

