Spatial and temporal control of targeting Polo-like kinase during meiotic prophase
- PMID: 32997737
- PMCID: PMC7594494
- DOI: 10.1083/jcb.202006094
Spatial and temporal control of targeting Polo-like kinase during meiotic prophase
Abstract
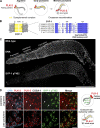
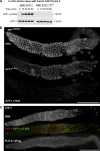
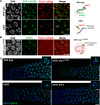
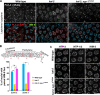
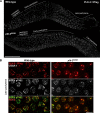
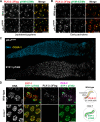
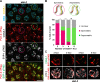
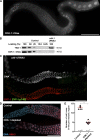
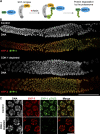
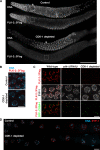
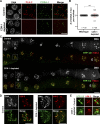
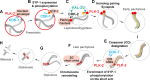
Polo-like kinases (PLKs) play widely conserved roles in orchestrating meiotic chromosome dynamics. However, how PLKs are targeted to distinct subcellular localizations during meiotic progression remains poorly understood. Here, we demonstrate that the cyclin-dependent kinase CDK-1 primes the recruitment of PLK-2 to the synaptonemal complex (SC) through phosphorylation of SYP-1 in C. elegans. SYP-1 phosphorylation by CDK-1 occurs just before meiotic onset. However, PLK-2 docking to the SC is prevented by the nucleoplasmic HAL-2/3 complex until crossover designation, which constrains PLK-2 to special chromosomal regions known as pairing centers to ensure proper homologue pairing and synapsis. PLK-2 is targeted to crossover sites primed by CDK-1 and spreads along the SC by reinforcing SYP-1 phosphorylation on one side of each crossover only when threshold levels of crossovers are generated. Thus, the integration of chromosome-autonomous signaling and a nucleus-wide crossover-counting mechanism partitions holocentric chromosomes relative to the crossover site, which ultimately defines the pattern of chromosome segregation during meiosis I.
© 2020 Brandt et al.
Figures

References
-
- Argunhan B., Leung W.-K., Afshar N., Terentyev Y., Subramanian V.V., Murayama Y., Hochwagen A., Iwasaki H., Tsubouchi T., and Tsubouchi H.. 2017. Fundamental cell cycle kinases collaborate to ensure timely destruction of the synaptonemal complex during meiosis. EMBO J. 36:2488–2509. 10.15252/embj.201695895 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

