Accuracy of serological testing for SARS-CoV-2 antibodies: First results of a large mixed-method evaluation study
- PMID: 32997812
- PMCID: PMC7537154
- DOI: 10.1111/all.14608
Accuracy of serological testing for SARS-CoV-2 antibodies: First results of a large mixed-method evaluation study
Abstract
Background: Serological immunoassays that can identify protective immunity against SARS-CoV-2 are needed to adapt quarantine measures, assess vaccination responses, and evaluate donor plasma. To date, however, the utility of such immunoassays remains unclear. In a mixed-design evaluation study, we compared the diagnostic accuracy of serological immunoassays that are based on various SARS-CoV-2 proteins and assessed the neutralizing activity of antibodies in patient sera.
Methods: Consecutive patients admitted with confirmed SARS-CoV-2 infection were prospectively followed alongside medical staff and biobank samples from winter 2018/2019. An in-house enzyme-linked immunosorbent assay utilizing recombinant receptor-binding domain (RBD) of the SARS-CoV-2 spike protein was developed and compared to three commercially available enzyme-linked immunosorbent assays (ELISAs) targeting the nucleoprotein (N), the S1 domain of the spike protein (S1), and a lateral flow immunoassay (LFI) based on full-length spike protein. Neutralization assays with live SARS-CoV-2 were performed.
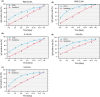
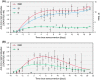
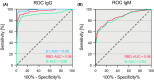
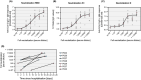
Results: One thousand four hundred and seventy-seven individuals were included comprising 112 SARS-CoV-2 positives (defined as a positive real-time PCR result; prevalence 7.6%). IgG seroconversion occurred between day 0 and day 21. While the ELISAs showed sensitivities of 88.4% for RBD, 89.3% for S1, and 72.9% for N protein, the specificity was above 94% for all tests. Out of 54 SARS-CoV-2 positive individuals, 96.3% showed full neutralization of live SARS-CoV-2 at serum dilutions ≥ 1:16, while none of the 6 SARS-CoV-2-negative sera revealed neutralizing activity.
Conclusions: ELISAs targeting RBD and S1 protein of SARS-CoV-2 are promising immunoassays which shall be further evaluated in studies verifying diagnostic accuracy and protective immunity against SARS-CoV-2.
Keywords: Antibodies; COVID-19 [Supplementary Concept]; COVID-19 diagnostic testing [Supplementary Concept]; Enzyme-Linked Immunosorbent Assay [Mesh]; Neutralizing [Mesh]; Severe Acute Respiratory Syndrome Coronavirus 2 [Supplementary Concept].
© 2020 The Authors. Allergy published by European Academy of Allergy and Clinical Immunology and John Wiley & Sons Ltd.
Conflict of interest statement
All authors declare that there is no conflict of interests.
Figures


References
-
- Krammer F, Simon V. Serology assays to manage COVID‐19. Science. 2020;368(6495):1060‐1061. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

