Modulation of the Apurinic/Apyrimidinic Endonuclease Activity of Human APE1 and of Its Natural Polymorphic Variants by Base Excision Repair Proteins
- PMID: 32998246
- PMCID: PMC7583023
- DOI: 10.3390/ijms21197147
Modulation of the Apurinic/Apyrimidinic Endonuclease Activity of Human APE1 and of Its Natural Polymorphic Variants by Base Excision Repair Proteins
Abstract
Human apurinic/apyrimidinic endonuclease 1 (APE1) is known to be a critical player of the base excision repair (BER) pathway. In general, BER involves consecutive actions of DNA glycosylases, AP endonucleases, DNA polymerases, and DNA ligases. It is known that these proteins interact with APE1 either at upstream or downstream steps of BER. Therefore, we may propose that even a minor disturbance of protein-protein interactions on the DNA template reduces coordination and repair efficiency. Here, the ability of various human DNA repair enzymes (such as DNA glycosylases OGG1, UNG2, and AAG; DNA polymerase Polβ; or accessory proteins XRCC1 and PCNA) to influence the activity of wild-type (WT) APE1 and its seven natural polymorphic variants (R221C, N222H, R237A, G241R, M270T, R274Q, and P311S) was tested. Förster resonance energy transfer-based kinetic analysis of abasic site cleavage in a model DNA substrate was conducted to detect the effects of interacting proteins on the activity of WT APE1 and its single-nucleotide polymorphism (SNP) variants. The results revealed that WT APE1 activity was stimulated by almost all tested DNA repair proteins. For the SNP variants, the matters were more complicated. Analysis of two SNP variants, R237A and G241R, suggested that a positive charge in this area of the APE1 surface impairs the protein-protein interactions. In contrast, variant R221C (where the affected residue is located near the DNA-binding site) showed permanently lower activation relative to WT APE1, whereas neighboring SNP N222H did not cause a noticeable difference as compared to WT APE1. Buried substitution P311S had an inconsistent effect, whereas each substitution at the DNA-binding site, M270T and R274Q, resulted in the lowest stimulation by BER proteins. Protein-protein molecular docking was performed between repair proteins to identify amino acid residues involved in their interactions. The data uncovered differences in the effects of BER proteins on APE1, indicating an important role of protein-protein interactions in the coordination of the repair pathway.
Keywords: AP endonuclease; DNA repair; coordination of DNA repair process; protein–protein interaction; single-nucleotide polymorphism.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
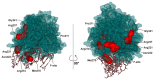

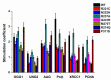
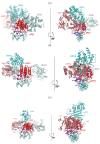


Similar articles
-
The impact of single-nucleotide polymorphisms of human apurinic/apyrimidinic endonuclease 1 on specific DNA binding and catalysis.Biochimie. 2019 Aug;163:73-83. doi: 10.1016/j.biochi.2019.05.015. Epub 2019 May 28. Biochimie. 2019. PMID: 31150756
-
The role of the N-terminal domain of human apurinic/apyrimidinic endonuclease 1, APE1, in DNA glycosylase stimulation.DNA Repair (Amst). 2018 Apr;64:10-25. doi: 10.1016/j.dnarep.2018.02.001. Epub 2018 Feb 11. DNA Repair (Amst). 2018. PMID: 29475157
-
Dynamic Processing of a Common Oxidative DNA Lesion by the First Two Enzymes of the Base Excision Repair Pathway.J Mol Biol. 2021 Mar 5;433(5):166811. doi: 10.1016/j.jmb.2021.166811. Epub 2021 Jan 13. J Mol Biol. 2021. PMID: 33450252 Free PMC article.
-
Single-molecule studies of repair proteins in base excision repair.BMB Rep. 2025 Jan;58(1):17-23. doi: 10.5483/BMBRep.2024-0178. BMB Rep. 2025. PMID: 39701025 Free PMC article. Review.
-
When DNA repair goes wrong: BER-generated DNA-protein crosslinks to oxidative lesions.DNA Repair (Amst). 2016 Aug;44:103-109. doi: 10.1016/j.dnarep.2016.05.014. Epub 2016 May 20. DNA Repair (Amst). 2016. PMID: 27264558 Free PMC article. Review.
Cited by
-
Detection of Uracil-Excising DNA Glycosylases in Cancer Cell Samples Using a Three-Dimensional DNAzyme Walker.ACS Meas Sci Au. 2024 May 8;4(4):459-466. doi: 10.1021/acsmeasuresciau.4c00011. eCollection 2024 Aug 21. ACS Meas Sci Au. 2024. PMID: 39184356 Free PMC article.
-
The Impact of Human DNA Glycosylases on the Activity of DNA Polymerase β toward Various Base Excision Repair Intermediates.Int J Mol Sci. 2023 May 31;24(11):9594. doi: 10.3390/ijms24119594. Int J Mol Sci. 2023. PMID: 37298543 Free PMC article.
-
Optimizing the Comet Assay-Based In Vitro DNA Repair Assay for Placental Tissue: A Pilot Study with Pre-Eclamptic Patients.Int J Mol Sci. 2023 Dec 22;25(1):187. doi: 10.3390/ijms25010187. Int J Mol Sci. 2023. PMID: 38203356 Free PMC article.
-
Position-Dependent Effects of AP Sites Within an hTERT Promoter G-Quadruplex Scaffold on Quadruplex Stability and Repair Activity of the APE1 Enzyme.Int J Mol Sci. 2025 Jan 2;26(1):337. doi: 10.3390/ijms26010337. Int J Mol Sci. 2025. PMID: 39796192 Free PMC article.
-
The base excision repair process: comparison between higher and lower eukaryotes.Cell Mol Life Sci. 2021 Dec;78(24):7943-7965. doi: 10.1007/s00018-021-03990-9. Epub 2021 Nov 3. Cell Mol Life Sci. 2021. PMID: 34734296 Free PMC article. Review.
References
-
- Lindahl T., Kubota Y., Klungland A. Reconstitution of the dna base excision-repair pathway with purified human enzymes. FASEB J. 1996;4:1069–1076.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

