Genome-Wide Identification of Barley ABC Genes and Their Expression in Response to Abiotic Stress Treatment
- PMID: 32998428
- PMCID: PMC7599588
- DOI: 10.3390/plants9101281
Genome-Wide Identification of Barley ABC Genes and Their Expression in Response to Abiotic Stress Treatment
Abstract
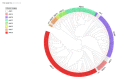
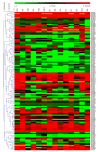
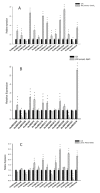
Adenosine triphosphate-binding cassette transporters (ABC transporters) participate in various plant growth and abiotic stress responses. In the present study, 131 ABC genes in barley were systematically identified using bioinformatics. Based on the classification method of the family in rice, these members were classified into eight subfamilies (ABCA-ABCG, ABCI). The conserved domain, amino acid composition, physicochemical properties, chromosome distribution, and tissue expression of these genes were predicted and analyzed. The results showed that the characteristic motifs of the barley ABC genes were highly conserved and there were great diversities in the homology of the transmembrane domain, the number of exons, amino acid length, and the molecular weight, whereas the span of the isoelectric point was small. Tissue expression profile analysis suggested that ABC genes possess non-tissue specificity. Ultimately, 15 differentially expressed genes exhibited diverse expression responses to stress treatments including drought, cadmium, and salt stress, indicating that the ABCB and ABCG subfamilies function in the response to abiotic stress in barley.
Keywords: ABC gene family; abiotic stress; barley; gene expression.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Identification of genomic ATP binding cassette (ABC) transporter genes and Cd-responsive ABCs in Brassica napus.Gene. 2018 Jul 20;664:139-151. doi: 10.1016/j.gene.2018.04.060. Epub 2018 Apr 27. Gene. 2018. PMID: 29709635
-
Genome-wide identification of Oryza sativa: A new insight for advanced analysis of ABC transporter genes associated with the degradation of four pesticides.Gene. 2022 Aug 5;834:146613. doi: 10.1016/j.gene.2022.146613. Epub 2022 May 25. Gene. 2022. PMID: 35643224
-
Genome-wide analysis of the ATP-binding cassette (ABC) transporter gene family in sea lamprey and Japanese lamprey.BMC Genomics. 2015 Jun 6;16(1):436. doi: 10.1186/s12864-015-1677-z. BMC Genomics. 2015. PMID: 26047617 Free PMC article.
-
Functions of ABC transporters in plants.Essays Biochem. 2011 Sep 7;50(1):145-60. doi: 10.1042/bse0500145. Essays Biochem. 2011. PMID: 21967056 Review.
-
Towards Identification of the Substrates of ATP-Binding Cassette Transporters.Plant Physiol. 2018 Sep;178(1):18-39. doi: 10.1104/pp.18.00325. Epub 2018 Jul 9. Plant Physiol. 2018. PMID: 29987003 Free PMC article. Review.
Cited by
-
Genome-Wide Identification and Analysis of the ABCF Gene Family in Triticum aestivum.Int J Mol Sci. 2023 Nov 18;24(22):16478. doi: 10.3390/ijms242216478. Int J Mol Sci. 2023. PMID: 38003668 Free PMC article.
-
Auxin Transporters-A Biochemical View.Cold Spring Harb Perspect Biol. 2022 Feb 1;14(2):a039875. doi: 10.1101/cshperspect.a039875. Cold Spring Harb Perspect Biol. 2022. PMID: 34127449 Free PMC article. Review.
-
Reprisal of Schima superba to Mn stress and exploration of its defense mechanism through transcriptomic analysis.Front Plant Sci. 2022 Oct 6;13:1022686. doi: 10.3389/fpls.2022.1022686. eCollection 2022. Front Plant Sci. 2022. PMID: 36311055 Free PMC article.
-
The maize ATP-binding cassette (ABC) transporter ZmMRPA6 confers cold and salt stress tolerance in plants.Plant Cell Rep. 2023 Dec 23;43(1):13. doi: 10.1007/s00299-023-03094-7. Plant Cell Rep. 2023. PMID: 38135780
-
Genome-Wide Identification and Expression Analysis of the Stearoyl-Acyl Carrier Protein Δ9 Desaturase Gene Family under Abiotic Stress in Barley.Int J Mol Sci. 2023 Dec 21;25(1):113. doi: 10.3390/ijms25010113. Int J Mol Sci. 2023. PMID: 38203283 Free PMC article.
References
-
- Lane T.S., Rempe C.S., Davitt J., Staton M.E., Peng Y., Soltis D.E., Melkonian M., Deyholos M., Leebens-Mack J.H., Chase M. Diversity of ABC transporter genes across the plant kingdom and their potential utility in biotechnology. BMC Biotechnol. 2017;16:47. doi: 10.1186/s12896-016-0277-6. - DOI - PMC - PubMed
-
- Liu Y.Q., Zhao Y.F. Structure and mechanism of ABC transporter. Chin. Bull. Life Sci. 2017;29:223–229.
Grants and funding
LinkOut - more resources
Full Text Sources

