Interferon-Induced Transmembrane Protein 3 Is a Virus-Associated Protein Which Suppresses Porcine Reproductive and Respiratory Syndrome Virus Replication by Blocking Viral Membrane Fusion
- PMID: 32999030
- PMCID: PMC7925183
- DOI: 10.1128/JVI.01350-20
Interferon-Induced Transmembrane Protein 3 Is a Virus-Associated Protein Which Suppresses Porcine Reproductive and Respiratory Syndrome Virus Replication by Blocking Viral Membrane Fusion
Abstract
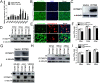
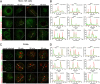
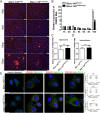
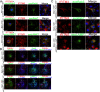
Porcine reproductive and respiratory syndrome virus (PRRSV) infection eliminates production of type I interferons (IFNs) in host cells, which triggers an antiviral immune response through the induction of downstream IFN-stimulated genes (ISGs), thus escaping the fate of host-mediated clearance. The IFN-induced transmembrane 3 (IFITM3) has recently been identified as an ISG and plays a pivotal role against enveloped RNA viruses by restricting cell entry. However, the role of IFITM3 in PRRSV replication is unknown. The present study demonstrated that overexpression of IFITM3 suppresses PRRSV replication, while silencing of endogenous IFITM3 prominently promoted PRRSV replication. Additionally, it was shown that IFITM3 undergoes S-palmitoylation and ubiquitination modification, and both posttranslational modifications contribute to the anti-PRRSV activity of IFITM3. Further study showed that PRRSV particles are transported into endosomes and then into lysosomes during the early stages of infection, and confocal microscopy results revealed that PRRSV particles are transported to IFITM3-positive cellular vesicles. By using a single virus particle fluorescent labeling technique, we confirmed that IFITM3 can restrict PRRSV membrane fusion by inducing accumulation of cholesterol in cellular vesicles. Additionally, we found that both endogenous and exogenous IFITM3 are incorporated into newly producing PRRS virions and diminish viral intrinsic infectivity. By using cell coculture systems, we found that IFITM3 effectively restricted PRRSV intercellular transmission, which may have been caused by disrupted membrane fusion and reduced viral infectivity. In conclusion, our results demonstrate, for the first time, that swine IFITM3 interferes with the life cycle of PRRSV, and possibly other enveloped arteritis viruses, at multiple steps.IMPORTANCE Porcine reproductive and respiratory syndrome (PRRS), which is caused by PRRS virus (PRRSV), is of great economic significance to the swine industry. Due to the complicated immune escape mechanisms of PRRSV, there are no effective vaccines or therapeutic drugs currently available against PRRS. Identification of cellular factors and underlying mechanisms that establish an effective antiviral state against PRRSV can provide unique strategies for developing antiviral vaccines or drugs. As an interferon (IFN)-stimulated gene, the role of IFN-induced transmembrane 3 (IFITM3) in PRRSV infection has not been reported as of yet. In the present study, it was shown that IFITM3 can exert a potent anti-PRRSV effect, and PRRS virions are trafficked to IFITM3-containing cell vesicles, where viral membrane fusion is impaired by cholesterol accumulation that is induced by IFITM3. Additionally, both endogenous and exogenous IFITM3 are incorporated into newly assembled progeny virions, and this decreased their intrinsic infectivity.
Keywords: IFITM3; PRRSV; cellular vesicles; cholesterol; membrane fusion.
Copyright © 2020 Zhang et al.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

