Ancestral function of Inhibitors-of-kappaB regulates Caenorhabditis elegans development
- PMID: 32999373
- PMCID: PMC7527347
- DOI: 10.1038/s41598-020-73146-5
Ancestral function of Inhibitors-of-kappaB regulates Caenorhabditis elegans development
Abstract
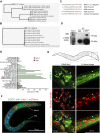
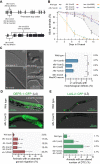
Mammalian IκB proteins (IκBs) exert their main function as negative regulators of NF-κB, a central signaling pathway controlling immunity and inflammation. An alternative chromatin role for IκBs has been shown to affect stemness and cell differentiation. However, the involvement of NF-κB in this function has not been excluded. NFKI-1 and IKB-1 are IκB homologs in Caenorhabditis elegans, which lacks NF-κB nuclear effectors. We found that nfki-1 and ikb-1 mutants display developmental defects that phenocopy mutations in Polycomb and UTX-1 histone demethylase, suggesting a role for C. elegans IκBs in chromatin regulation. Further supporting this possibility (1) we detected NFKI-1 in the nucleus of cells; (2) NFKI-1 and IKB-1 bind to histones and Polycomb proteins, (3) and associate with chromatin in vivo, and (4) mutations in nfki-1 and ikb-1 alter chromatin marks. Based on these results, we propose that ancestral IκB inhibitors modulate Polycomb activity at specific gene subsets with an impact on development.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

