Automated identification of leukocyte subsets improves standardization of database-guided expert-supervised diagnostic orientation in acute leukemia: a EuroFlow study
- PMID: 32999413
- PMCID: PMC7806506
- DOI: 10.1038/s41379-020-00677-7
Automated identification of leukocyte subsets improves standardization of database-guided expert-supervised diagnostic orientation in acute leukemia: a EuroFlow study
Abstract
Precise classification of acute leukemia (AL) is crucial for adequate treatment. EuroFlow has previously designed an AL orientation tube (ALOT) to guide toward the relevant classification panel and final diagnosis. In this study, we designed and validated an algorithm for automated (database-supported) gating and identification (AGI tool) of cell subsets within samples stained with ALOT. A reference database of normal peripheral blood (PB, n = 41) and bone marrow (BM; n = 45) samples analyzed with the ALOT was constructed, and served as a reference for the AGI tool to automatically identify normal cells. Populations not unequivocally identified as normal cells were labeled as checks and were classified by an expert. Additional normal BM (n = 25) and PB (n = 43) and leukemic samples (n = 109), analyzed in parallel by experts and the AGI tool, were used to evaluate the AGI tool. Analysis of normal PB and BM samples showed low percentages of checks (<3% in PB, <10% in BM), with variations between different laboratories. Manual analysis and AGI analysis of normal and leukemic samples showed high levels of correlation between cell numbers (r2 > 0.95 for all cell types in PB and r2 > 0.75 in BM) and resulted in highly concordant classification of leukemic cells by our previously published automated database-guided expert-supervised orientation tool for immunophenotypic diagnosis and classification of acute leukemia (Compass tool). Similar data were obtained using alternative, commercially available tubes, confirming the robustness of the developed tools. The AGI tool represents an innovative step in minimizing human intervention and requirements in expertise, toward a "sample-in and result-out" approach which may result in more objective and reproducible data analysis and diagnostics. The AGI tool may improve quality of immunophenotyping in individual laboratories, since high percentages of checks in normal samples are an alert on the quality of the internal procedures.
Conflict of interest statement
JJMvD, AO, JFM, VHJvdV, LL, EM, and TS each report being one of the inventors on the EuroFlow-owned patent PCT/NL2010/050332 (Methods, reagents and kits for flowcytometric immunophenotyping of normal, reactive, and malignant leukocytes). The Infinicyt software is based on intellectual property (IP) of some EuroFlow laboratories (University of Salamanca in Spain and Federal University of Rio de Janeiro in Brazil) and the scientific input of other EuroFlow members. All above mentioned intellectual property and related patents are licensed to Cytognos (Salamanca, ES) and BD Biosciences (San José, CA), which companies pay royalties to the EuroFlow Consortium. These royalties are exclusively used for continuation of the EuroFlow collaboration and sustainability of the EuroFlow consortium. MB reports to belong to speakers bureau/honoraria (Amgen, Celgene, Janssen), Advisory board/committee (Amgen, Janssen), to be involved in consultancy (Amgen, Incyte, PRMA) and to receive research funding (Affimed, Amgen, Celgene, Regeneron). VHJvdV reports a Laboratory Services Agreement with BD Biosciences; all related fees are for the Erasmus MC. JJMvD and AO report an Educational Services Agreement from BD Biosciences (San José, CA) and a Scientific Advisor Agreement with Cytognos; all related fees and honoraria are for the involved university departments at Leiden University Medical Center and University of Salamanca. GG, SB, AHD, and RF are employees of Cytognos. The other authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




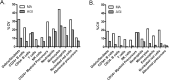
References
-
- Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al. editors. WHO classification of tumours of haematopoietic and lymphoid tissues. Revised 4th ed. Lyon: International Agency for Research on Cancer; 2017. p. 10–13.
-
- Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al. editors. WHO classification of tumours of haematopoietic and lymphoid tissues. Revised 4th ed. Lyon: International Agency for Research on Cancer; 2017. p. 199–213.
-
- Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al. editors. WHO classification of tumours of haematopoietic and lymphoid tissues. Revised 4th ed. Lyon: International Agency for Research on Cancer; 2017. p. 129–71.
-
- Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al. editors. WHO classification of tumours of haematopoietic and lymphoid tissues. Revised 4th ed. Lyon: International Agency for Research on Cancer; 2017. p. 179–87.

