Direct interactions with commensal streptococci modify intercellular communication behaviors of Streptococcus mutans
- PMID: 32999420
- PMCID: PMC8027600
- DOI: 10.1038/s41396-020-00789-7
Direct interactions with commensal streptococci modify intercellular communication behaviors of Streptococcus mutans
Abstract
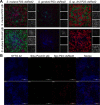
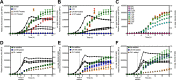
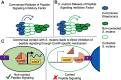
The formation of dental caries is a complex process that ultimately leads to damage of the tooth enamel from acids produced by microbes in attached biofilms. The bacterial interactions occurring within these biofilms between cariogenic bacteria, such as the mutans streptococci, and health-associated commensal streptococci, are thought to be critical determinants of health and disease. To better understand these interactions, a Streptococcus mutans reporter strain that actively monitors cell-cell communication via peptide signaling was cocultured with different commensal streptococci. Signaling by S. mutans, normally highly active in monoculture, was completely inhibited by several species of commensals, but only when the bacteria were in direct contact with S. mutans. We identified a novel gene expression pattern that occurred in S. mutans when cultured directly with these commensals. Finally, mutant derivatives of commensals lacking previously shown antagonistic gene products displayed wild-type levels of signal inhibition in cocultures. Collectively, these results reveal a novel pathway(s) in multiple health-associated commensal streptococci that blocks peptide signaling and induces a common contact-dependent pattern of differential gene expression in S. mutans. Understanding the molecular basis for this inhibition will assist in the rational design of new risk assessments, diagnostics, and treatments for the most pervasive oral infectious diseases.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





Similar articles
-
Growth with Commensal Streptococci Alters Streptococcus mutans Behaviors.J Dent Res. 2023 Apr;102(4):450-458. doi: 10.1177/00220345221145906. Epub 2023 Jan 23. J Dent Res. 2023. PMID: 36688378 Free PMC article.
-
Human saliva modifies growth, biofilm architecture, and competitive behaviors of oral streptococci.mSphere. 2024 Feb 28;9(2):e0077123. doi: 10.1128/msphere.00771-23. Epub 2024 Feb 6. mSphere. 2024. PMID: 38319113 Free PMC article.
-
Amino Sugars Modify Antagonistic Interactions between Commensal Oral Streptococci and Streptococcus mutans.Appl Environ Microbiol. 2019 May 2;85(10):e00370-19. doi: 10.1128/AEM.00370-19. Print 2019 May 15. Appl Environ Microbiol. 2019. PMID: 30877119 Free PMC article.
-
Competitive dynamics and balance between Streptococcus mutans and commensal streptococci in oral microecology.Crit Rev Microbiol. 2025 May;51(3):532-543. doi: 10.1080/1040841X.2024.2389386. Epub 2024 Aug 12. Crit Rev Microbiol. 2025. PMID: 39132685 Review.
-
[Streptococcus mutans and oral streptococci in dental plaque].Can J Microbiol. 2011 Jan;57(1):1-20. doi: 10.1139/w10-095. Can J Microbiol. 2011. PMID: 21217792 Review. French.
Cited by
-
Impact of different oral treatments on the composition of the supragingival plaque microbiome.J Oral Microbiol. 2022 Oct 31;14(1):2138251. doi: 10.1080/20002297.2022.2138251. eCollection 2022. J Oral Microbiol. 2022. PMID: 36338832 Free PMC article.
-
Growth with Commensal Streptococci Alters Streptococcus mutans Behaviors.J Dent Res. 2023 Apr;102(4):450-458. doi: 10.1177/00220345221145906. Epub 2023 Jan 23. J Dent Res. 2023. PMID: 36688378 Free PMC article.
-
Social networking at the microbiome-host interface.Infect Immun. 2023 Sep 14;91(9):e0012423. doi: 10.1128/iai.00124-23. Epub 2023 Aug 18. Infect Immun. 2023. PMID: 37594277 Free PMC article. Review.
-
Zinc import mediated by AdcABC is critical for colonization of the dental biofilm by Streptococcus mutans in an animal model.Mol Oral Microbiol. 2021 Jun;36(3):214-224. doi: 10.1111/omi.12337. Epub 2021 May 10. Mol Oral Microbiol. 2021. PMID: 33819383 Free PMC article.
-
Complete Genome Sequence of Streptococcus oralis 34.Microbiol Resour Announc. 2021 Sep 2;10(35):e0076021. doi: 10.1128/MRA.00760-21. Epub 2021 Sep 2. Microbiol Resour Announc. 2021. PMID: 34472981 Free PMC article.
References
-
- Ghoul M, Mitri S. The ecology and evolution of microbial competition. Trends Microbiol. 2016;24:833–45. - PubMed
-
- Granato ET, Meiller-Legrand TA, Foster KR. The evolution and ecology of bacterial warfare. Curr Biol. 2019;29:R521–37. - PubMed
-
- Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–46. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

