Two-step method for isolation of high-quality RNA from stored seeds of maize rich in starch
- PMID: 32999811
- PMCID: PMC7494719
- DOI: 10.1007/s13205-020-02424-w
Two-step method for isolation of high-quality RNA from stored seeds of maize rich in starch
Abstract
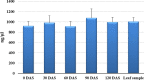
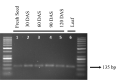
A modified SDS-Trizol method was optimized for isolation of total RNA from the stored maize seeds at regular interval of one month for 4 months. Use of SDS extraction buffer before the use of Trizol reduced the co-precipitation problem associated with high carbohydrate content in the seed. Recorded mean RNA yield from seeds across the storage intervals was 978.6 ± 65.46 ng/µl. Average spectrophotometric values (A 260/280) of isolated RNA varied from 1.974 ± 0.033 to 1.998 ± 0.022. Attempts to isolate RNA from green leaves using Trizol method also ensured comparable quality and quantity of the isolated RNA. RNA yield from fresh leaves was recorded 1008.2 ± 77.088 ng/µl which is slightly higher than the mean RNA yield from seeds across months. Observed mean A 260/280 values of isolated RNA were 1.984 ± 0.030. DNase treatment further improved the A 260/280 ratio in both seeds (2.003 ± 0.006) and leaves (2.012 ± 0.037). High quality and quantity along with integrity of the isolated RNA was ensured through downstream analysis after RNA extraction such as first-strand cDNA synthesis and normal PCR. Extraction of RNA from the stored seeds using modified SDS-based Trizol method and from fresh leaves using Trizol method opened new possibility of understanding role of key genes involving developmental steps especially in the stored seeds.
Keywords: Dry condition; Leaf; RNA; Seed; Starch; Storage.
© King Abdulaziz City for Science and Technology 2020.
Conflict of interest statement
Conflict of interestOn behalf of all authors, the corresponding author states that there is no conflict of interest. The authors declare that they have no conflict of interests.
Figures


Similar articles
-
Isolation of high quality RNA from cereal seeds containing high levels of starch.Phytochem Anal. 2012 Mar-Apr;23(2):159-63. doi: 10.1002/pca.1337. Epub 2011 Jul 8. Phytochem Anal. 2012. PMID: 21739496
-
A universal method for high-quality RNA extraction from plant tissues rich in starch, proteins and fiber.Sci Rep. 2020 Oct 9;10(1):16887. doi: 10.1038/s41598-020-73958-5. Sci Rep. 2020. PMID: 33037299 Free PMC article.
-
Extractions of High Quality RNA from the Seeds of Jerusalem Artichoke and Other Plant Species with High Levels of Starch and Lipid.Plants (Basel). 2013 Apr 29;2(2):302-16. doi: 10.3390/plants2020302. Plants (Basel). 2013. PMID: 27137377 Free PMC article.
-
Efficient recovery of proteins from multiple source samples after TRIzol(®) or TRIzol(®)LS RNA extraction and long-term storage.BMC Genomics. 2013 Mar 15;14:181. doi: 10.1186/1471-2164-14-181. BMC Genomics. 2013. PMID: 23496794 Free PMC article.
-
MicroRNA isolation and stability in stored RNA samples.Biochem Biophys Res Commun. 2009 Dec 4;390(1):1-4. doi: 10.1016/j.bbrc.2009.09.061. Epub 2009 Sep 19. Biochem Biophys Res Commun. 2009. PMID: 19769940 Review.
Cited by
-
Exploring novel SNPs and candidate genes associated with seed allometry in Pisum sativum L.Physiol Mol Biol Plants. 2024 Sep;30(9):1449-1462. doi: 10.1007/s12298-024-01499-6. Epub 2024 Aug 19. Physiol Mol Biol Plants. 2024. PMID: 39310699
-
Transcriptomic Analysis of Cold-Induced Temporary Cysts in Marine Dinoflagellate Prorocentrum cordatum.Int J Mol Sci. 2025 Jun 6;26(12):5432. doi: 10.3390/ijms26125432. Int J Mol Sci. 2025. PMID: 40564896 Free PMC article.
-
Transcriptomic response of marine dinoflagellate Prorocentrum cordatum to phosphorus deficiency.Sci Rep. 2025 May 29;15(1):18797. doi: 10.1038/s41598-025-02014-x. Sci Rep. 2025. PMID: 40442192 Free PMC article.
-
Expression analysis of β-carotene hydroxylase1 and opaque2 genes governing accumulation of provitamin-A, lysine and tryptophan during kernel development in biofortified sweet corn.3 Biotech. 2021 Jul;11(7):325. doi: 10.1007/s13205-021-02837-1. Epub 2021 Jun 11. 3 Biotech. 2021. PMID: 34194909 Free PMC article.
-
Harnessing the potential of millets for climate-resilient and sustainable agriculture.Front Plant Sci. 2025 Jun 11;16:1574699. doi: 10.3389/fpls.2025.1574699. eCollection 2025. Front Plant Sci. 2025. PMID: 40567416 Free PMC article. Review.
References
-
- Ahlfen SV, Schlumpberger M (2010) Effects of low A260/A230 ratios in RNA preparations on downstream applications. Qiagen Gene Exp Newslett 15
-
- Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation: version II. Plant Mol Biol Rep. 1983;1:19–21. doi: 10.1007/BF02712670. - DOI
-
- Delobel C, Lacher S, Mazzara M, Van Den Eede G. Event specific method for quantification of maize line Bt11 using real time PCR. Eur Comm Jt Res Centre. 2008 doi: 10.2788/4370. - DOI
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

