AbSeq Protocol Using the Nano-Well Cartridge-Based Rhapsody Platform to Generate Protein and Transcript Expression Data on the Single-Cell Level
- PMID: 33000001
- PMCID: PMC7523635
- DOI: 10.1016/j.xpro.2020.100092
AbSeq Protocol Using the Nano-Well Cartridge-Based Rhapsody Platform to Generate Protein and Transcript Expression Data on the Single-Cell Level
Abstract
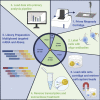
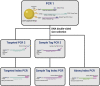
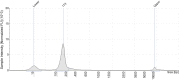
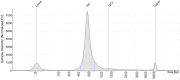
By including oligonucleotide-labeled antibodies into high-throughput single-cell RNA-sequencing protocols, combined transcript and protein expression data can be acquired on the single-cell level. Here, we describe a protocol for the combined analysis of over 40 proteins and 400 genes on over 104 cells using the nano-well based Rhapsody platform. We also include a workflow for sample multiplexing, which uniquely identifies the initial source of cells (such as tissue type or donor) in the downstream analysis after upstream pooling. For complete information on the use and execution of this protocol, please refer to Mair et al. (2020).
Conflict of interest statement
DECLARATION OF INTERESTS J.R.E., F.M. G.B., and M.P. declare no competing interests. J.M., A.J.T., S.M., and M.N. are employees of BD Biosciences.
Figures






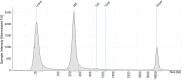

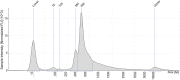
References
-
- Andersen M.N., Al-Karradi S.N., Kragstrup T.W., Hokland M. Elimination of erroneous results in flow cytometry caused by antibody binding to Fc receptors on human monocytes and macrophages. Cytometry A. 2016;89:1001–1009. - PubMed
-
- Azimifar S.B., Nagaraj N., Cox J., Mann M. Cell-type-resolved quantitative proteomics of murine liver. Cell Metab. 2014;20:1076–1087. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

