Miniaturization of Smart-seq2 for Single-Cell and Single-Nucleus RNA Sequencing
- PMID: 33000004
- PMCID: PMC7501729
- DOI: 10.1016/j.xpro.2020.100081
Miniaturization of Smart-seq2 for Single-Cell and Single-Nucleus RNA Sequencing
Abstract
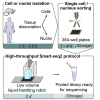
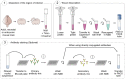
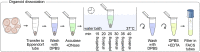
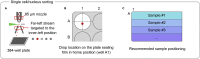
This protocol presents a plate-based workflow to perform RNA sequencing analysis of single cells/nuclei using Smart-seq2. We describe (1) the dissociation procedures for cell/nucleus isolation from the mouse brain and human organoids, (2) the flow sorting of single cells/nuclei into 384-well plates, and (3) the preparation of libraries following miniaturization of the Smart-seq2 protocol using a liquid-handling robot. This pipeline allows for the reliable, high-throughput, and cost-effective preparation of mouse and human samples for full-length deep single-cell/nucleus RNA sequencing. For complete details on the use and execution of this protocol, please refer to Bowers et al. (2020).
© 2020 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Collection of cells for single-cell RNA sequencing using high-resolution fluorescence microscopy.STAR Protoc. 2021 Aug 5;2(3):100718. doi: 10.1016/j.xpro.2021.100718. eCollection 2021 Sep 17. STAR Protoc. 2021. PMID: 34401784 Free PMC article.
-
Full-Length Single-Cell RNA-Sequencing with FLASH-seq.Methods Mol Biol. 2023;2584:123-164. doi: 10.1007/978-1-0716-2756-3_5. Methods Mol Biol. 2023. PMID: 36495447
-
Isolation and RNA sequencing of single nuclei from Drosophila tissues.STAR Protoc. 2022 May 20;3(2):101417. doi: 10.1016/j.xpro.2022.101417. eCollection 2022 Jun 17. STAR Protoc. 2022. PMID: 35620068 Free PMC article.
-
Review of Single-Cell RNA Sequencing in the Heart.Int J Mol Sci. 2020 Nov 6;21(21):8345. doi: 10.3390/ijms21218345. Int J Mol Sci. 2020. PMID: 33172208 Free PMC article. Review.
-
Single-cell RNA and protein profiling of immune cells from the mouse brain and its border tissues.Nat Protoc. 2022 Oct;17(10):2354-2388. doi: 10.1038/s41596-022-00716-4. Epub 2022 Aug 5. Nat Protoc. 2022. PMID: 35931780 Review.
Cited by
-
Visualization of individual cell division history in complex tissues using iCOUNT.Cell Stem Cell. 2021 Nov 4;28(11):2020-2034.e12. doi: 10.1016/j.stem.2021.08.012. Epub 2021 Sep 14. Cell Stem Cell. 2021. PMID: 34525348 Free PMC article.
-
Microglia contribute to the postnatal development of cortical somatostatin-positive inhibitory cells and to whisker-evoked cortical activity.Cell Rep. 2022 Aug 16;40(7):111209. doi: 10.1016/j.celrep.2022.111209. Cell Rep. 2022. PMID: 35977514 Free PMC article.
-
Long-read RNA sequencing of transposable elements from single cells using CELLO-seq.Nat Protoc. 2025 Jul 16:10.1038/s41596-025-01203-2. doi: 10.1038/s41596-025-01203-2. Online ahead of print. Nat Protoc. 2025. PMID: 40670609 Free PMC article. Review.
-
FASN-dependent de novo lipogenesis is required for brain development.Proc Natl Acad Sci U S A. 2022 Jan 11;119(2):e2112040119. doi: 10.1073/pnas.2112040119. Proc Natl Acad Sci U S A. 2022. PMID: 34996870 Free PMC article.
-
Transcriptional profiling of mouse peripheral nerves to the single-cell level to build a sciatic nerve ATlas (SNAT).Elife. 2021 Apr 23;10:e58591. doi: 10.7554/eLife.58591. Elife. 2021. PMID: 33890853 Free PMC article.
References
-
- Bowers M., Liang T., Gonzalez-Bohorquez D., Zocher S., Jaeger B.N., Kovacs W.J., Rohrl C., Cramb K.M.L., Winterer J., Kruse M., Dimitrieva S., Overall R.W. FASN-dependent lipid metabolism links neurogenic stem/progenitor cell activity to learning and memory deficits. Cell Stem Cell. 2020;27:98–109.e11. - PubMed
-
- Camp J.G., Badsha F., Florio M., Kanton S., Gerber T., Wilsch-Brauninger M., Lewitus E., Sykes A., Hevers W., Lancaster M., Knoblich J.A., Lachmann R. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc. Natl. Acad. Sci. U S A. 2015;112:15672–15677. - PMC - PubMed
-
- Hempel C.M., Sugino K., Nelson S.B. A manual method for the purification of fluorescently labeled neurons from the mammalian brain. Nat. Protoc. 2007;2:2924–2929. - PubMed
-
- Jaeger B.N., Linker S.B., Parylak S.L., Barron J.J., Gallina I.S., Saavedra C.D., Fitzpatrick C., Lim C.K., Schafer S.T., Lacar B., Jessberger S., Gage F.H. A novel environment-evoked transcriptional signature predicts reactivity in single dentate granule neurons. Nat. Commun. 2018;9:3084. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

