Alteration of enzymes and their application to nucleic acid amplification (Review)
- PMID: 33000189
- PMCID: PMC7521554
- DOI: 10.3892/ijmm.2020.4726
Alteration of enzymes and their application to nucleic acid amplification (Review)
Abstract
Since the discovery of polymerase chain reaction (PCR) in 1985, several methods have been developed to achieve nucleic acid amplification, and are currently used in various fields including clinical diagnosis and life science research. Thus, a wealth of information has accumulated regarding nucleic acid‑related enzymes. In this review, some nucleic acid‑related enzymes were selected and the recent advances in their modification along with their application to nucleic acid amplification were described. The discussion also focused on optimization of the corresponding reaction conditions. Using newly developed enzymes under well‑optimized reaction conditions, the sensitivity, specificity, and fidelity of nucleic acid tests can be improved successfully.
Figures

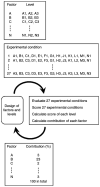
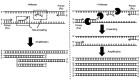

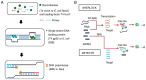
Similar articles
-
Enzyme-Assisted Nucleic Acid Amplification in Molecular Diagnosis: A Review.Biosensors (Basel). 2023 Jan 19;13(2):160. doi: 10.3390/bios13020160. Biosensors (Basel). 2023. PMID: 36831926 Free PMC article. Review.
-
Exponential Isothermal Amplification of Nucleic Acids and Assays for Proteins, Cells, Small Molecules, and Enzyme Activities: An EXPAR Example.Angew Chem Int Ed Engl. 2018 Sep 10;57(37):11856-11866. doi: 10.1002/anie.201712217. Epub 2018 Aug 15. Angew Chem Int Ed Engl. 2018. PMID: 29704305 Review.
-
Nucleic acid isothermal amplification technologies: a review.Nucleosides Nucleotides Nucleic Acids. 2008 Mar;27(3):224-43. doi: 10.1080/15257770701845204. Nucleosides Nucleotides Nucleic Acids. 2008. PMID: 18260008 Review.
-
Advances in nucleic acid-based detection methods.Clin Microbiol Rev. 1992 Oct;5(4):370-86. doi: 10.1128/CMR.5.4.370. Clin Microbiol Rev. 1992. PMID: 1423216 Free PMC article. Review.
-
Nucleic acid amplification free biosensors for pathogen detection.Biosens Bioelectron. 2020 Apr 1;153:112049. doi: 10.1016/j.bios.2020.112049. Epub 2020 Jan 27. Biosens Bioelectron. 2020. PMID: 32056663 Review.
Cited by
-
A Guide to Using FASTPCR Software for PCR, In Silico PCR, and Oligonucleotide Analysis.Methods Mol Biol. 2022;2392:223-243. doi: 10.1007/978-1-0716-1799-1_16. Methods Mol Biol. 2022. PMID: 34773626
-
Characterization and PCR Application of Family B DNA Polymerases from Thermococcus stetteri.Life (Basel). 2024 Nov 25;14(12):1544. doi: 10.3390/life14121544. Life (Basel). 2024. PMID: 39768253 Free PMC article.
-
RT-qPCR Detection of SARS-CoV-2: No Need for a Dedicated Reverse Transcription Step.Int J Mol Sci. 2022 Jan 24;23(3):1303. doi: 10.3390/ijms23031303. Int J Mol Sci. 2022. PMID: 35163227 Free PMC article.
-
Optimization of reaction condition of recombinase polymerase amplification to detect SARS-CoV-2 DNA and RNA using a statistical method.Biochem Biophys Res Commun. 2021 Aug 27;567:195-200. doi: 10.1016/j.bbrc.2021.06.023. Epub 2021 Jun 10. Biochem Biophys Res Commun. 2021. PMID: 34166918 Free PMC article.
-
Loop-Mediated Isothermal Amplification Detection of SARS-CoV-2 and Myriad Other Applications.J Biomol Tech. 2021 Sep;32(3):228-275. doi: 10.7171/jbt.21-3203-017. J Biomol Tech. 2021. PMID: 35136384 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

