Investigating aberrantly expressed microRNAs in peripheral blood mononuclear cells from patients with treatment‑resistant schizophrenia using miRNA sequencing and integrated bioinformatics
- PMID: 33000265
- PMCID: PMC7533444
- DOI: 10.3892/mmr.2020.11513
Investigating aberrantly expressed microRNAs in peripheral blood mononuclear cells from patients with treatment‑resistant schizophrenia using miRNA sequencing and integrated bioinformatics
Abstract
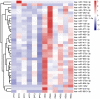
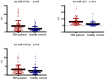
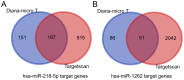
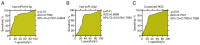
Treatment‑resistant schizophrenia (TRS) is a common phenotype of schizophrenia that places a considerable burden on patients as well as on society. TRS is known for its tendency to relapse and uncontrollable nature, with a poor response to antipsychotics other than clozapine. Therefore, it is urgent to identify objective biological markers, so as to guide its treatment and associated clinical work. In the present study, the peripheral blood mononuclear cells (PBMCs) of patients with TRS and a healthy control group, which were gender‑, age‑ and ethnicity‑matched, were subjected to microRNA (miRNA/miR) sequencing to screen out the top three miRNAs with the highest fold change values. These were then validated in the TRS (n=34) and healthy control (n=31) groups by reverse transcription‑quantitative PCR. For two of the top three miRNAs, the PCR results were in accordance with the sequencing result (P<0.01), while the third miRNA exhibited the opposite trend (P<0.01). To elucidate the functions of these two miRNAs, Homo sapiens (hsa)‑miR‑218‑5p and hsa‑miR‑1262 and their regulatory network, target gene prediction was first performed using online TargetScan and Diana‑micro T software. Bioinformatics analysis was then performed using functional enrichment analysis to determine the Gene Ontology terms in the category biological process and the Kyoto Encyclopedia of Genes and Genomes pathways. It was revealed that these target genes were markedly associated with the nervous system and brain function, and it was obvious that the differentially expressed miRNAs most likely participated in the pathogenesis of TRS. A receiver operating characteristic curve was generated to confirm the distinct diagnostic value of these two miRNAs. It was concluded that aberrantly expressed miRNAs in PMBCs may be implicated in the pathogenesis of TRS and may serve as specific peripheral blood‑based biomarkers for the early diagnosis of TRS.
Figures

Similar articles
-
Altered microRNA Expression in Peripheral Blood Mononuclear Cells from Young Patients with Schizophrenia.J Mol Neurosci. 2015 Jul;56(3):562-71. doi: 10.1007/s12031-015-0503-z. Epub 2015 Feb 11. J Mol Neurosci. 2015. PMID: 25665552
-
Identification of miR-22-3p, miR-92a-3p, and miR-137 in peripheral blood as biomarker for schizophrenia.Psychiatry Res. 2018 Jul;265:70-76. doi: 10.1016/j.psychres.2018.03.080. Epub 2018 Apr 7. Psychiatry Res. 2018. PMID: 29684772
-
Bioinformatic Analysis of Potential microRNAs in Ischemic Stroke.J Stroke Cerebrovasc Dis. 2016 Jul;25(7):1753-1759. doi: 10.1016/j.jstrokecerebrovasdis.2016.03.023. Epub 2016 Apr 14. J Stroke Cerebrovasc Dis. 2016. PMID: 27151415
-
Peripheral blood MicroRNAs as biomarkers of schizophrenia: expectations from a meta-analysis that combines deep learning methods.World J Biol Psychiatry. 2024 Jan-Feb;25(1):65-81. doi: 10.1080/15622975.2023.2258975. Epub 2023 Sep 13. World J Biol Psychiatry. 2024. PMID: 37703215 Review.
-
The Promise of Predictive Biomarkers for Antipsychotic Efficacy: A Review of Peripheral microRNAs to Evaluate Schizophrenia Treatment Response.Prim Care Companion CNS Disord. 2022 Apr 7;24(2):21nr03048. doi: 10.4088/PCC.21nr03048. Prim Care Companion CNS Disord. 2022. PMID: 35395147 Review.
Cited by
-
Peripheral biomarkers of treatment-resistant schizophrenia: Genetic, inflammation and stress perspectives.Front Pharmacol. 2022 Oct 12;13:1005702. doi: 10.3389/fphar.2022.1005702. eCollection 2022. Front Pharmacol. 2022. PMID: 36313375 Free PMC article. Review.
-
The Role of microRNA in Schizophrenia: A Scoping Review.Int J Mol Sci. 2024 Jul 12;25(14):7673. doi: 10.3390/ijms25147673. Int J Mol Sci. 2024. PMID: 39062916 Free PMC article.
-
miR-218-2 regulates cognitive functions in the hippocampus through complement component 3-dependent modulation of synaptic vesicle release.Proc Natl Acad Sci U S A. 2021 Apr 6;118(14):e2021770118. doi: 10.1073/pnas.2021770118. Proc Natl Acad Sci U S A. 2021. PMID: 33782126 Free PMC article.
-
Extracellular microRNAs associated with psychiatric symptoms in the Normative Aging Study.J Psychiatr Res. 2024 Oct;178:270-277. doi: 10.1016/j.jpsychires.2024.08.017. Epub 2024 Aug 15. J Psychiatr Res. 2024. PMID: 39173451 Free PMC article.
-
Recent advancements in biomarker research in schizophrenia: mapping the road from bench to bedside.Metab Brain Dis. 2022 Oct;37(7):2197-2211. doi: 10.1007/s11011-022-00926-5. Epub 2022 Mar 3. Metab Brain Dis. 2022. PMID: 35239143 Review.
References
-
- Lehman AF, Lieberman JA, Dixon LB, McGlashan TH, Miller AL, Perkins DO, Kreyenbuhl J, American Psychiatric Association; Steering Committee on Practice Guidelines Practice guideline for the treatment of patients with schizophrenia, second edition. Am J Psychiatry. 2004;161:1–56. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

