Electroblotting through a tryptic membrane for LC-MS/MS analysis of proteins separated in electrophoretic gels
- PMID: 33000802
- PMCID: PMC7704035
- DOI: 10.1039/d0an01380c
Electroblotting through a tryptic membrane for LC-MS/MS analysis of proteins separated in electrophoretic gels
Abstract
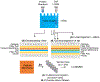
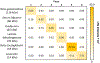
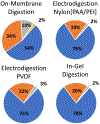
Digestion of proteins separated via sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) remains a popular method for protein identification using mass-spectrometry based proteomics. Although robust and routine, the in-gel digestion procedure is laborious and time-consuming. Electroblotting to a capture membrane prior to digestion reduces preparation steps but requires on-membrane digestion that yields fewer peptides than in-gel digestion. This paper develops direct electroblotting through a trypsin-containing membrane to a capture membrane to simplify extraction and digestion of proteins separated by SDS-PAGE. Subsequent liquid chromatography-tandem mass spectrometry (LC-MS/MS) identifies the extracted peptides. Analysis of peptides from different capture membrane pieces shows that electrodigestion does not greatly disturb the spatial resolution of a standard protein mixture separated by SDS-PAGE. Electrodigestion of an Escherichia coli (E. coli) cell lysate requires four hours of total sample preparation and results in only 13% fewer protein identifications than in-gel digestion, which can take 24 h. Compared to simple electroblotting and protein digestion on a poly(vinylidene difluoride) (PVDF) capture membrane, adding a trypsin membrane to the electroblot increases the number of protein identifications by 22%. Additionally, electrodigestion experiments using capture membranes coated with polyelectrolyte layers identify a higher fraction of small proteolytic peptides than capture on PVDF or in-gel digestion.
Conflict of interest statement
Conflicts of interest
There are no conflicts to declare.
Figures




Similar articles
-
High yield electroblotting onto polyvinylidene difluoride membranes from polyacrylamide gels.Electrophoresis. 1992 Jan-Feb;13(1-2):59-64. doi: 10.1002/elps.1150130112. Electrophoresis. 1992. PMID: 1587256
-
BAC-DROP: Rapid Digestion of Proteome Fractionated via Dissolvable Polyacrylamide Gel Electrophoresis and Its Application to Bottom-Up Proteomics Workflow.J Proteome Res. 2021 Mar 5;20(3):1535-1543. doi: 10.1021/acs.jproteome.0c00749. Epub 2020 Dec 24. J Proteome Res. 2021. PMID: 33356312
-
Tube-gel digestion: a novel proteomic approach for high throughput analysis of membrane proteins.Mol Cell Proteomics. 2005 Dec;4(12):1948-58. doi: 10.1074/mcp.M500138-MCP200. Epub 2005 Sep 8. Mol Cell Proteomics. 2005. PMID: 16150870 Free PMC article.
-
Protein Extraction from Gels: A Brief Review.Methods Mol Biol. 2019;1855:479-482. doi: 10.1007/978-1-4939-8793-1_40. Methods Mol Biol. 2019. PMID: 30426441 Review.
-
Extraction of proteins from gels: a brief review.Methods Mol Biol. 2012;869:403-5. doi: 10.1007/978-1-61779-821-4_33. Methods Mol Biol. 2012. PMID: 22585504 Free PMC article. Review.
Cited by
-
In-Membrane Enrichment and Peptic Digestion to Facilitate Analysis of Monoclonal Antibody Glycosylation.Anal Chem. 2024 Apr 23;96(16):6347-6355. doi: 10.1021/acs.analchem.4c00030. Epub 2024 Apr 12. Anal Chem. 2024. PMID: 38607313 Free PMC article.
-
Electroblotting through Enzymatic Membranes to Enhance Molecular Tissue Imaging.J Am Soc Mass Spectrom. 2021 Jul 7;32(7):1689-1699. doi: 10.1021/jasms.1c00046. Epub 2021 Jun 10. J Am Soc Mass Spectrom. 2021. PMID: 34110793 Free PMC article.
-
Potential of the Novel Slot Blot Method with a PVDF Membrane for Protein Identification and Quantification in Kampo Medicines.Membranes (Basel). 2023 Dec 1;13(12):896. doi: 10.3390/membranes13120896. Membranes (Basel). 2023. PMID: 38132900 Free PMC article.
-
Slot Blot- and Electrospray Ionization-Mass Spectrometry/Matrix-Assisted Laser Desorption/Ionization-Mass Spectrometry-Based Novel Analysis Methods for the Identification and Quantification of Advanced Glycation End-Products in the Urine.Int J Mol Sci. 2024 Sep 5;25(17):9632. doi: 10.3390/ijms25179632. Int J Mol Sci. 2024. PMID: 39273579 Free PMC article.
References
-
- Thorsen SF, Gromova I, Christensen IJ, Fredriksson S, Andersen CL, Nielsen HJ, Stenvang J, Moreira JMA, Gel-Based Proteomics of Clinical Samples Identifies Potential Serological Biomarkers for Early Detection of Colorectal Cancer. Int. J. Mol. Sci 2019, 20(23), 6082 DOI: 10.3390/ijms20236082 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

