Mechanisms of epitranscriptomic gene regulation
- PMID: 33001446
- PMCID: PMC7855325
- DOI: 10.1002/bip.23403
Mechanisms of epitranscriptomic gene regulation
Abstract
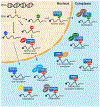
Chemical modifications on RNA can regulate fundamental biological processes. Recent efforts have illuminated the chemical diversity of posttranscriptional ("epitranscriptomic") modifications on eukaryotic messenger RNA and have begun to elucidate their biological roles. In this review, we discuss our current molecular understanding of epitranscriptomic RNA modifications and their effects on gene expression. In particular, we highlight the role of modifications in mediating RNA-protein interactions, RNA structure, and RNA-RNA base pairing, and how these macromolecular interactions control biological processes in the cell.
© 2020 Wiley Periodicals LLC.
Conflict of interest statement
CONFLICT OF INTEREST
No competing interests have been declared.
Figures
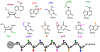
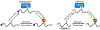
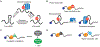
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

