Microbial and transcriptional differences elucidate atopic dermatitis heterogeneity across skin sites
- PMID: 33001460
- PMCID: PMC8246754
- DOI: 10.1111/all.14606
Microbial and transcriptional differences elucidate atopic dermatitis heterogeneity across skin sites
Abstract
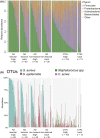
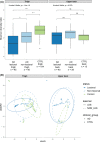
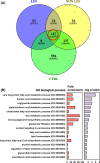
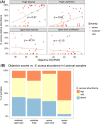
It is well established that different sites in healthy human skin are colonized by distinct microbial communities due to different physiological conditions. However, few studies have explored microbial heterogeneity between skin sites in diseased skin, such as atopic dermatitis (AD) lesions. To address this issue, we carried out deep analysis of the microbiome and transcriptome in the skin of a large cohort of AD patients and healthy volunteers, comparing two physiologically different sites: upper back and posterior thigh. Microbiome samples and biopsies were obtained from both lesional and nonlesional skin to identify changes related to the disease process. Transcriptome analysis revealed distinct disease-related gene expression profiles depending on anatomical location, with keratinization dominating the transcriptomic signatures in posterior thigh, and lipid metabolism in the upper back. Moreover, we show that relative abundance of Staphylococcus aureus is associated with disease severity in the posterior thigh, but not in the upper back. Our results suggest that AD may select for similar microbes in different anatomical locations-an "AD-like microbiome," but distinct microbial dynamics can still be observed when comparing posterior thigh to upper back. This study highlights the importance of considering the variability across skin sites when studying the development of skin inflammation.
Keywords: atopic dermatitis; inflammation; microbiome.
© 2020 The Authors. Allergy published by European Academy of Allergy and Clinical Immunology and John Wiley & Sons Ltd.
Conflict of interest statement
Dr Ottman reports grants from BIOMAP IMI2 821511, during the conduct of the study; Dr Barrientos‐Somarribas has nothing to disclose; Dr Fyhrquist has nothing to disclose; Dr Alexander has nothing to disclose; Dr Wisgrill has nothing to disclose; Dr Olah has nothing to disclose; Dr Tsoka has nothing to disclose; Dr Greco has nothing to disclose; Dr Levi‐Schaffer has nothing to disclose; Dr Soumelis has nothing to disclose; Dr Schröder has nothing to disclose; Dr Kere has nothing to disclose; Dr Nestle reports other from Sanofi, outside the submitted work; Dr Barker has nothing to disclose; Dr Ranki reports grants from EU FP7/2007‐2013, during the conduct of the study; Dr Lauerma reports grants from Orion Corporation, outside the submitted work; Dr Homey reports grants from EU‐MAARS, grants from EU‐BIOMAP, grants from DFG‐FOR2690‐HO 2092/7‐1, during the conduct of the study; grants and personal fees from Galderma, personal fees from AbbVie, personal fees from Janssen, personal fees from Sanofi/Regeneron, personal fees from Leo Pharmaceuticals, outside the submitted work; Dr Andersson has nothing to disclose; and Dr Alenius reports grants from BIOMAP IMI2 821511, during the conduct of the study.
Figures




References
-
- Byrd AL, Belkaid Y, Segre JA. The human skin microbiome. Nat Rev Microbiol. 2018;16:143‐155. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

