Large-scale variation in single nucleotide polymorphism density within the laboratory axolotl (Ambystoma mexicanum)
- PMID: 33001517
- PMCID: PMC8715502
- DOI: 10.1002/dvdy.257
Large-scale variation in single nucleotide polymorphism density within the laboratory axolotl (Ambystoma mexicanum)
Abstract
Background: Recent efforts to assemble and analyze the Ambystoma mexicanum genome have dramatically improved the potential to develop molecular tools and pursue genome-wide analyses of genetic variation.
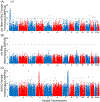
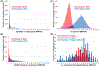
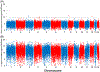
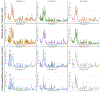
Results: To better resolve the distribution and origins of genetic variation with A mexicanum, we compared DNA sequence data for two laboratory A mexicanum and one A tigrinum to identify 702 million high confidence polymorphisms distributed across the 32 Gb genome. While the wild-caught A tigrinum was generally more polymorphic in a genome-wide sense, several multi-megabase regions were identified from A mexicanum genomes that were actually more polymorphic than A tigrinum. Analysis of polymorphism and repeat content reveals that these regions likely originated from the intentional hybridization of A mexicanum and A tigrinum that was used to introduce the albino mutation into laboratory stocks.
Conclusions: Our findings show that axolotl genomes are variable with respect to introgressed DNA from a highly polymorphic species. It seems likely that other divergent regions will be discovered with additional sequencing of A mexicanum. This has practical implications for designing molecular probes and suggests a need to study A mexicanum phenotypic variation and genome evolution across the tiger salamander clade.
Keywords: SNPs; axolotl; genome; hybrid; salamander.
© 2020 Wiley Periodicals LLC.
Figures










References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

