In utero human intestine harbors unique metabolome, including bacterial metabolites
- PMID: 33001863
- PMCID: PMC7710283
- DOI: 10.1172/jci.insight.138751
In utero human intestine harbors unique metabolome, including bacterial metabolites
Abstract
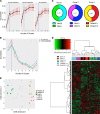
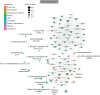
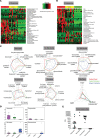
Symbiotic microbial colonization through the establishment of the intestinal microbiome is critical to many intestinal functions, including nutrient metabolism, intestinal barrier integrity, and immune regulation. Recent studies suggest that education of intestinal immunity may be ongoing in utero. However, the drivers of this process are unknown. The microbiome and its byproducts are one potential source. Whether a fetal intestinal microbiome exists is controversial, and whether microbially derived metabolites are present in utero is unknown. Here, we aimed to determine whether bacterial DNA and microbially derived metabolites can be detected in second trimester human intestinal samples. Although we were unable to amplify bacterial DNA from fetal intestines, we report a fetal metabolomic intestinal profile with an abundance of bacterially derived and host-derived metabolites commonly produced in response to microbiota. Though we did not directly assess their source and function, we hypothesize that these microbial-associated metabolites either come from the maternal microbiome and are vertically transmitted to the fetus to prime the fetal immune system and prepare the gastrointestinal tract for postnatal microbial encounters or are produced locally by bacteria that were below our detection threshold.
Keywords: Gastroenterology; Intermediary metabolism; Metabolism.
Conflict of interest statement
ex
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

