RNAthor - fast, accurate normalization, visualization and statistical analysis of RNA probing data resolved by capillary electrophoresis
- PMID: 33002005
- PMCID: PMC7529196
- DOI: 10.1371/journal.pone.0239287
RNAthor - fast, accurate normalization, visualization and statistical analysis of RNA probing data resolved by capillary electrophoresis
Abstract
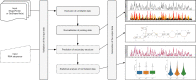
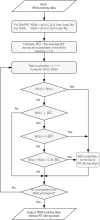
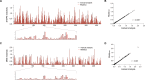
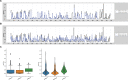
RNAs adopt specific structures to perform their functions, which are critical to fundamental cellular processes. For decades, these structures have been determined and modeled with strong support from computational methods. Still, the accuracy of the latter ones depends on the availability of experimental data, for example, chemical probing information that can define pseudo-energy constraints for RNA folding algorithms. At the same time, diverse computational tools have been developed to facilitate analysis and visualization of data from RNA structure probing experiments followed by capillary electrophoresis or next-generation sequencing. RNAthor, a new software tool for the fully automated normalization of SHAPE and DMS probing data resolved by capillary electrophoresis, has recently joined this collection. RNAthor automatically identifies unreliable probing data. It normalizes the reactivity information to a uniform scale and uses it in the RNA secondary structure prediction. Our web server also provides tools for fast and easy RNA probing data visualization and statistical analysis that facilitates the comparison of multiple data sets. RNAthor is freely available at http://rnathor.cs.put.poznan.pl/.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
RNAProbe: a web server for normalization and analysis of RNA structure probing data.Nucleic Acids Res. 2020 Jul 2;48(W1):W292-W299. doi: 10.1093/nar/gkaa396. Nucleic Acids Res. 2020. PMID: 32504492 Free PMC article.
-
Automated band annotation for RNA structure probing experiments with numerous capillary electrophoresis profiles.Bioinformatics. 2015 Sep 1;31(17):2808-15. doi: 10.1093/bioinformatics/btv282. Epub 2015 May 5. Bioinformatics. 2015. PMID: 25943472 Free PMC article.
-
Automated RNA 3D Structure Prediction with RNAComposer.Methods Mol Biol. 2016;1490:199-215. doi: 10.1007/978-1-4939-6433-8_13. Methods Mol Biol. 2016. PMID: 27665601
-
Computational analysis of RNA structures with chemical probing data.Methods. 2015 Jun;79-80:60-6. doi: 10.1016/j.ymeth.2015.02.003. Epub 2015 Feb 14. Methods. 2015. PMID: 25687190 Free PMC article. Review.
-
Energy-based RNA consensus secondary structure prediction in multiple sequence alignments.Methods Mol Biol. 2014;1097:125-41. doi: 10.1007/978-1-62703-709-9_7. Methods Mol Biol. 2014. PMID: 24639158 Review.
Cited by
-
RNA Chemical Labeling with Site-Specific, Relative Quantification by Mass Spectrometry for the Structural Study of a Neomycin-Sensing Riboswitch Aptamer Domain.Chempluschem. 2022 Nov;87(11):e202200256. doi: 10.1002/cplu.202200256. Epub 2022 Oct 11. Chempluschem. 2022. PMID: 36220343 Free PMC article.
-
Computational Pipeline for Reference-Free Comparative Analysis of RNA 3D Structures Applied to SARS-CoV-2 UTR Models.Int J Mol Sci. 2022 Aug 25;23(17):9630. doi: 10.3390/ijms23179630. Int J Mol Sci. 2022. PMID: 36077037 Free PMC article.
-
Evaluation of the stereochemical quality of predicted RNA 3D models in the RNA-Puzzles submissions.RNA. 2022 Feb;28(2):250-262. doi: 10.1261/rna.078685.121. Epub 2021 Nov 24. RNA. 2022. PMID: 34819324 Free PMC article.
-
Machine learning for RNA 2D structure prediction benchmarked on experimental data.Brief Bioinform. 2023 May 19;24(3):bbad153. doi: 10.1093/bib/bbad153. Brief Bioinform. 2023. PMID: 37096592 Free PMC article. Review.
-
Transformers in RNA structure prediction: A review.Comput Struct Biotechnol J. 2025 Mar 17;27:1187-1203. doi: 10.1016/j.csbj.2025.03.021. eCollection 2025. Comput Struct Biotechnol J. 2025. PMID: 40213272 Free PMC article. Review.
References
-
- Waterman MS, Smith TF (1978) Rna Secondary Structure—Complete Mathematical-Analysis. Mathematical Biosciences 42: 257–266.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

