Transcriptome analysis of non human primate-induced pluripotent stem cell-derived cardiomyocytes in 2D monolayer culture vs. 3D engineered heart tissue
- PMID: 33002105
- PMCID: PMC8318103
- DOI: 10.1093/cvr/cvaa281
Transcriptome analysis of non human primate-induced pluripotent stem cell-derived cardiomyocytes in 2D monolayer culture vs. 3D engineered heart tissue
Abstract
Aims: Stem cell therapy has shown promise for treating myocardial infarction via re-muscularization and paracrine signalling in both small and large animals. Non-human primates (NHPs), such as rhesus macaques (Macaca mulatta), are primarily utilized in preclinical trials due to their similarity to humans, both genetically and physiologically. Currently, induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs) are delivered into the infarcted myocardium by either direct cell injection or an engineered tissue patch. Although both approaches have advantages in terms of sample preparation, cell-host interaction, and engraftment, how the iPSC-CMs respond to ischaemic conditions in the infarcted heart under these two different delivery approaches remains unclear. Here, we aim to gain a better understanding of the effects of hypoxia on iPSC-CMs at the transcriptome level.
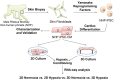
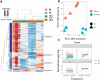
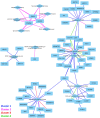
Methods and results: NHP iPSC-CMs in both monolayer culture (2D) and engineered heart tissue (EHT) (3D) format were exposed to hypoxic conditions to serve as surrogates of direct cell injection and tissue implantation in vivo, respectively. Outcomes were compared at the transcriptome level. We found the 3D EHT model was more sensitive to ischaemic conditions and similar to the native in vivo myocardium in terms of cell-extracellular matrix/cell-cell interactions, energy metabolism, and paracrine signalling.
Conclusion: By exposing NHP iPSC-CMs to different culture conditions, transcriptome profiling improves our understanding of the mechanism of ischaemic injury.
Keywords: Cardiomyocytes; Engineered heart tissue; Hypoxia; Induced pluripotent stem cells; Non-human primate; Transcriptome.
Published on behalf of the European Society of Cardiology. All rights reserved. © The Author(s) 2020. For permissions, please email: journals.permissions@oup.com.
Figures



Comment in
-
Three-dimensional unity of engineered heart tissue mimics the heart better than two-dimensional cellular diversity.Cardiovasc Res. 2021 Jul 27;117(9):1995-1997. doi: 10.1093/cvr/cvab052. Cardiovasc Res. 2021. PMID: 33580250 Free PMC article. No abstract available.
Similar articles
-
Comparison of Non-human Primate versus Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes for Treatment of Myocardial Infarction.Stem Cell Reports. 2018 Feb 13;10(2):422-435. doi: 10.1016/j.stemcr.2018.01.002. Epub 2018 Feb 1. Stem Cell Reports. 2018. PMID: 29398480 Free PMC article.
-
Combined Treatment of Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes and Endothelial Cells Regenerate the Infarcted Heart in Mice and Non-Human Primates.Circulation. 2023 Oct 31;148(18):1395-1409. doi: 10.1161/CIRCULATIONAHA.122.061736. Epub 2023 Sep 21. Circulation. 2023. PMID: 37732466 Free PMC article.
-
Advancing Human iPSC-Derived Cardiomyocyte Hypoxia Resistance for Cardiac Regenerative Therapies through a Systematic Assessment of In Vitro Conditioning.Int J Mol Sci. 2024 Sep 5;25(17):9627. doi: 10.3390/ijms25179627. Int J Mol Sci. 2024. PMID: 39273573 Free PMC article.
-
Maturation strategies and limitations of induced pluripotent stem cell-derived cardiomyocytes.Biosci Rep. 2021 Jun 25;41(6):BSR20200833. doi: 10.1042/BSR20200833. Biosci Rep. 2021. PMID: 33057659 Free PMC article. Review.
-
Induced pluripotent stem cell derived cardiac models: effects of Thymosin β4.Expert Opin Biol Ther. 2018 Jul;18(sup1):111-120. doi: 10.1080/14712598.2018.1473370. Expert Opin Biol Ther. 2018. PMID: 30063852 Review.
Cited by
-
Contractility and Calcium Transient Maturation in the Human iPSC-Derived Cardiac Microfibers.ACS Appl Mater Interfaces. 2022 Aug 10;14(31):35376-35388. doi: 10.1021/acsami.2c07326. Epub 2022 Jul 28. ACS Appl Mater Interfaces. 2022. PMID: 35901275 Free PMC article.
-
Bioengineering Systems for Modulating Notch Signaling in Cardiovascular Development, Disease, and Regeneration.J Cardiovasc Dev Dis. 2021 Sep 30;8(10):125. doi: 10.3390/jcdd8100125. J Cardiovasc Dev Dis. 2021. PMID: 34677194 Free PMC article. Review.
-
Bioprinting approaches in cardiac tissue engineering to reproduce blood-pumping heart function.iScience. 2024 Dec 20;28(1):111664. doi: 10.1016/j.isci.2024.111664. eCollection 2025 Jan 17. iScience. 2024. PMID: 39868032 Free PMC article. Review.
-
Retinal Ganglion Cell Replacement in Glaucoma Therapy: A Narrative Review.J Clin Med. 2024 Nov 27;13(23):7204. doi: 10.3390/jcm13237204. J Clin Med. 2024. PMID: 39685661 Free PMC article. Review.
-
Reconstructing the heart using iPSCs: Engineering strategies and applications.J Mol Cell Cardiol. 2021 Aug;157:56-65. doi: 10.1016/j.yjmcc.2021.04.006. Epub 2021 Apr 22. J Mol Cell Cardiol. 2021. PMID: 33895197 Free PMC article. Review.
References
-
- Anderson JL, Morrow DA.. Acute myocardial infarction. N Engl J Med 2017;376:2053–2064. - PubMed
-
- Benjamin EJ, Blaha MJ, Chiuve SE, Cushman M, Das SR, Deo R, de Ferranti SD, Floyd J, Fornage M, Gillespie C, Isasi CR, Jimenez MC, Jordan LC, Judd SE, Lackland D, Lichtman JH, Lisabeth L, Liu S, Longenecker CT, Mackey RH, Matsushita K, Mozaffarian D, Mussolino ME, Nasir K, Neumar RW, Palaniappan L, Pandey DK, Thiagarajan RR, Reeves MJ, Ritchey M, Rodriguez CJ, Roth GA, Rosamond WD, Sasson C, Towfighi A, Tsao CW, Turner MB, Virani SS, Voeks JH, Willey JZ, Wilkins JT, Wu JH, Alger HM, Wong SS, Muntner P.. American Heart Association Statistics C and Stroke Statistics S. Heart disease and stroke statistics-2017 update: a report from the American Heart Association. Circulation 2017;135:e146–e603. - PMC - PubMed
-
- Chong JJ, Yang X, Don CW, Minami E, Liu YW, Weyers JJ, Mahoney WM, Van Biber B, Cook SM, Palpant NJ, Gantz JA, Fugate JA, Muskheli V, Gough GM, Vogel KW, Astley CA, Hotchkiss CE, Baldessari A, Pabon L, Reinecke H, Gill EA, Nelson V, Kiem HP, Laflamme MA, Murry CE.. Human embryonic-stem-cell-derived cardiomyocytes regenerate non-human primate hearts. Nature 2014;510:273–277. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
- American Heart Association
- L30 HL138771/HL/NHLBI NIH HHS/United States
- R01 HL141371/HL/NHLBI NIH HHS/United States
- RT3-07798/California Institute of Regenerative Medicine
- S10 OD010417/OD/NIH HHS/United States
- K01 HL130608/HL/NHLBI NIH HHS/United States
- UG3 TR002588/TR/NCATS NIH HHS/United States
- California National Primate Research Center
- 18POST34030106/Postdoctoral Fellowship Award
- R01 HL145676/HL/NHLBI NIH HHS/United States
- R01 HL141851/HL/NHLBI NIH HHS/United States
- P51 OD011107/OD/NIH HHS/United States
- R25 HL145817/HL/NHLBI NIH HHS/United States
- Steven M. Gootter Foundation
- R24 HL085794/HL/NHLBI NIH HHS/United States
- T32 HL007444/HL/NHLBI NIH HHS/United States

