NusG-dependent RNA polymerase pausing is a frequent function of this universally conserved transcription elongation factor
- PMID: 33003953
- PMCID: PMC9404317
- DOI: 10.1080/10409238.2020.1828261
NusG-dependent RNA polymerase pausing is a frequent function of this universally conserved transcription elongation factor
Abstract
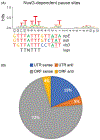
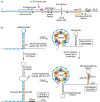
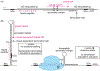
Although transcription by RNA polymerase (RNAP) is highly processive, elongation can be transiently halted by RNAP pausing. Pausing provides time for diverse regulatory events to occur such as RNA folding and regulatory factor binding. The transcription elongation factors NusA and NusG dramatically affect the frequency and duration of RNAP pausing, and hence regulation of transcription. NusG is the only transcription factor conserved in all three domains of life; its homolog in archaea and eukaryotes is Spt5. This review focuses on NusG-dependent pausing, which is a common occurrence in Bacillus subtilis. B. NusG induces pausing about once per 3 kb at a consensus TTNTTT motif in the non-template DNA strand within the paused transcription bubble. A conserved region of NusG contacts the TTNTTT motif to stabilize the paused transcription elongation complex (TEC) in multiple catalytically inactive RNAP conformations. The density of NusG-dependent pause sites is 3-fold higher in untranslated regions, suggesting that pausing could regulate the expression of hundreds of genes in B. subtilis. We describe how pausing in 5' leader regions contributes to regulating the expression of B. subtilis genes by transcription attenuation and translation control mechanisms. As opposed to the broadly accepted view that NusG is an anti-pausing factor, phylogenetic analyses suggest that NusG-dependent pausing is a widespread mechanism in bacteria. This function of NusG is consistent with the well-established role of its eukaryotic homolog Spt5 in promoter-proximal pausing. Since NusG is present in all domains of life, NusG-dependent pausing could be a conserved mechanism in all organisms.
Keywords: NusG; RNA polymerase pausing; Spt5; gene regulation; riboswitch; transcription attenuation; translation control.
Conflict of interest statement
Disclosure statement
The authors declare that this review was written in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Antson AA, Dodson EJ, Dodson G, Greaves RB, Chen X, Gollnick P. 1999. Structure of the trp RNA-binding attenuation protein, TRAP, bound to RNA. Nature. 401(6750): 235–242. - PubMed
-
- Antson AA, Otridge J, Brzozowski AM, Dodson EJ, Dodson GG, Wilson KS, Smith TM, Yang M, Kurecki T, Gollnick P. 1995. The structure of trp RNA-binding attenuation protein. Nature. 374(6524):693–700. - PubMed
-
- Artsimovitch I, Landick R. 2002. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell. 109(2):193–203. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
