Salt tolerance involved candidate genes in rice: an integrative meta-analysis approach
- PMID: 33004003
- PMCID: PMC7528482
- DOI: 10.1186/s12870-020-02679-8
Salt tolerance involved candidate genes in rice: an integrative meta-analysis approach
Abstract
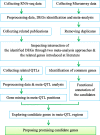
Background: Salinity, as one of the main abiotic stresses, critically threatens growth and fertility of main food crops including rice in the world. To get insight into the molecular mechanisms by which tolerant genotypes responds to the salinity stress, we propose an integrative meta-analysis approach to find the key genes involved in salinity tolerance. Herein, a genome-wide meta-analysis, using microarray and RNA-seq data was conducted which resulted in the identification of differentially expressed genes (DEGs) under salinity stress at tolerant rice genotypes. DEGs were then confirmed by meta-QTL analysis and literature review.
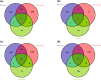
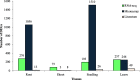
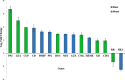
Results: A total of 3449 DEGs were detected in 46 meta-QTL positions, among which 1286, 86, 1729 and 348 DEGs were observed in root, shoot, seedling, and leaves tissues, respectively. Moreover, functional annotation of DEGs located in the meta-QTLs suggested some involved biological processes (e.g., ion transport, regulation of transcription, cell wall organization and modification as well as response to stress) and molecular function terms (e.g., transporter activity, transcription factor activity and oxidoreductase activity). Remarkably, 23 potential candidate genes were detected in Saltol and hotspot-regions overlying original QTLs for both yield components and ion homeostasis traits; among which, there were many unreported salinity-responsive genes. Some promising candidate genes were detected such as pectinesterase, peroxidase, transcription regulator, high-affinity potassium transporter, cell wall organization, protein serine/threonine phosphatase, and CBS domain cotaining protein.
Conclusions: The obtained results indicated that, the salt tolerant genotypes use qualified mechanisms particularly in sensing and signalling of the salt stress, regulation of transcription, ionic homeostasis, and Reactive Oxygen Species (ROS) scavenging in response to the salt stress.
Keywords: Meta- analysis; Microarray; Oryza sativa; QTLs; RNA-seq; Salinity stress.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Integrated RNA-Seq Analysis and Meta-QTLs Mapping Provide Insights into Cold Stress Response in Rice Seedling Roots.Int J Mol Sci. 2020 Jun 29;21(13):4615. doi: 10.3390/ijms21134615. Int J Mol Sci. 2020. PMID: 32610550 Free PMC article.
-
Mapping QTLs for traits related to salinity tolerance at seedling stage of rice (Oryza sativa L.): an agrigenomics study of an Iranian rice population.OMICS. 2013 May;17(5):242-51. doi: 10.1089/omi.2012.0097. OMICS. 2013. PMID: 23638881
-
Transcription dynamics of Saltol QTL localized genes encoding transcription factors, reveals their differential regulation in contrasting genotypes of rice.Funct Integr Genomics. 2017 Jan;17(1):69-83. doi: 10.1007/s10142-016-0529-5. Epub 2016 Nov 15. Funct Integr Genomics. 2017. PMID: 27848097
-
Salinity stress tolerance and omics approaches: revisiting the progress and achievements in major cereal crops.Heredity (Edinb). 2022 Jun;128(6):497-518. doi: 10.1038/s41437-022-00516-2. Epub 2022 Mar 5. Heredity (Edinb). 2022. PMID: 35249098 Free PMC article. Review.
-
Salt tolerance in rice: seedling and reproductive stage QTL mapping come of age.Theor Appl Genet. 2021 Nov;134(11):3495-3533. doi: 10.1007/s00122-021-03890-3. Epub 2021 Jul 21. Theor Appl Genet. 2021. PMID: 34287681 Free PMC article. Review.
Cited by
-
Association of a Specific OsCULLIN3c Haplotype with Salt Stress Responses in Local Thai Rice.Int J Mol Sci. 2024 Jan 15;25(2):1040. doi: 10.3390/ijms25021040. Int J Mol Sci. 2024. PMID: 38256116 Free PMC article.
-
QTLs and Genes for Salt Stress Tolerance: A Journey from Seed to Seed Continued.Plants (Basel). 2024 Apr 14;13(8):1099. doi: 10.3390/plants13081099. Plants (Basel). 2024. PMID: 38674508 Free PMC article. Review.
-
Molecular Alchemy: Converting Stress into Resilience via Secondary Metabolites and Calcium Signaling in Rice.Rice (N Y). 2025 May 5;18(1):32. doi: 10.1186/s12284-025-00783-7. Rice (N Y). 2025. PMID: 40325258 Free PMC article. Review.
-
Meta-analysis of identified genomic regions and candidate genes underlying salinity tolerance in rice (Oryza sativa L.).Sci Rep. 2024 Mar 8;14(1):5730. doi: 10.1038/s41598-024-54764-9. Sci Rep. 2024. PMID: 38459066 Free PMC article.
-
Development of a Temperate Climate-Adapted indica Multi-stress Tolerant Rice Variety by Pyramiding Quantitative Trait Loci.Rice (N Y). 2022 Apr 9;15(1):22. doi: 10.1186/s12284-022-00568-2. Rice (N Y). 2022. PMID: 35397732 Free PMC article.
References
-
- ZHANG H, et al. Progress of potato staple food research and industry development in China. J Integr Agric. 2017;16(12):2924–2932. doi: 10.1016/S2095-3119(17)61736-2. - DOI
-
- Hoang T, et al. Improvement of salinity stress tolerance in rice: challenges and opportunities. Agronomy. 2016;6(4):54. doi: 10.3390/agronomy6040054. - DOI
-
- Zeng L, Shannon MC, Lesch SM. Timing of salinity stress affects rice growth and yield components. Agric Water Manag. 2001;48(3):191–206. doi: 10.1016/S0378-3774(00)00146-3. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

