Dynamics and clinical relevance of maternal mRNA clearance during the oocyte-to-embryo transition in humans
- PMID: 33004802
- PMCID: PMC7530992
- DOI: 10.1038/s41467-020-18680-6
Dynamics and clinical relevance of maternal mRNA clearance during the oocyte-to-embryo transition in humans
Abstract
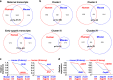
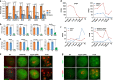
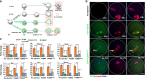
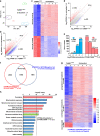
Maternal mRNA clearance is an essential process that occurs during maternal-to-zygotic transition (MZT). However, the dynamics, functional importance, and pathological relevance of maternal mRNA decay in human preimplantation embryos have not yet been analyzed. Here we report the zygotic genome activation (ZGA)-dependent and -independent maternal mRNA clearance processes during human MZT and demonstrate that subgroups of human maternal transcripts are sequentially removed by maternal (M)- and zygotic (Z)-decay pathways before and after ZGA. Key factors regulating M-decay and Z-decay pathways in mouse have similar expression pattern during human MZT, suggesting that YAP1-TEAD4 transcription activators, TUT4/7-mediated mRNA 3'-oligouridylation, and BTG4/CCR4-NOT-induced mRNA deadenylation may also be involved in the regulation of human maternal mRNA stability. Decreased expression of these factors and abnormal accumulation of maternal transcripts are observed in the development-arrested embryos of patients who seek assisted reproduction. Defects of M-decay and Z-decay are detected with high incidence in embryos that are arrested at the zygote and 8-cell stages, respectively. In addition, M-decay is not found to be affected by maternal TUBB8 mutations, although these mutations cause meiotic cell division defects and zygotic arrest, which indicates that mRNA decay is regulated independent of meiotic spindle assembly. Considering the correlations between maternal mRNA decay defects and early developmental arrest of in vitro fertilized human embryos, M-decay and Z-decay pathway activities may contribute to the developmental potential of human preimplantation embryos.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

