Analyzing synchronized clusters in neuron networks
- PMID: 33004897
- PMCID: PMC7530773
- DOI: 10.1038/s41598-020-73269-9
Analyzing synchronized clusters in neuron networks
Abstract
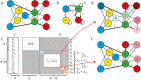
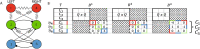
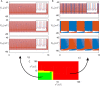
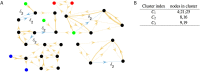
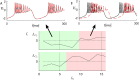
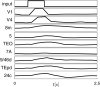
The presence of synchronized clusters in neuron networks is a hallmark of information transmission and processing. Common approaches to study cluster synchronization in networks of coupled oscillators ground on simplifying assumptions, which often neglect key biological features of neuron networks. Here we propose a general framework to study presence and stability of synchronous clusters in more realistic models of neuron networks, characterized by the presence of delays, different kinds of neurons and synapses. Application of this framework to two examples with different size and features (the directed network of the macaque cerebral cortex and the swim central pattern generator of a mollusc) provides an interpretation key to explain known functional mechanisms emerging from the combination of anatomy and neuron dynamics. The cluster synchronization analysis is carried out also by changing parameters and studying bifurcations. Despite some modeling simplifications in one of the examples, the obtained results are in good agreement with previously reported biological data.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

