Exploring celiac disease candidate pathways by global gene expression profiling and gene network cluster analysis
- PMID: 33004927
- PMCID: PMC7529771
- DOI: 10.1038/s41598-020-73288-6
Exploring celiac disease candidate pathways by global gene expression profiling and gene network cluster analysis
Abstract
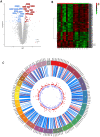
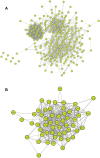
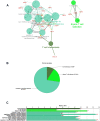
Celiac disease (CeD) is a gastrointestinal autoimmune disorder, whose specific molecular basis is not yet fully interpreted. Therefore, in this study, we compared the global gene expression profile of duodenum tissues from CeD patients, both at the time of disease diagnosis and after two years of the gluten-free diet. A series of advanced systems biology approaches like differential gene expression, protein-protein interactions, gene network-cluster analysis were deployed to annotate the candidate pathways relevant to CeD pathogenesis. The duodenum tissues from CeD patients revealed the differential expression of 106 up- and 193 down-regulated genes. The pathway enrichment of differentially expressed genes (DEGs) highlights the involvement of biological pathways related to loss of cell division regulation (cell cycle, p53 signalling pathway), immune system processes (NOD-like receptor signalling pathway, Th1, and Th2 cell differentiation, IL-17 signalling pathway) and impaired metabolism and absorption (mineral and vitamin absorptions and drug metabolism) in celiac disease. The molecular dysfunctions of these 3 biological events tend to increase the number of intraepithelial lymphocytes (IELs) and villous atrophy of the duodenal mucosa promoting the development of CeD. For the first time, this study highlights the involvement of aberrant cell division, immune system, absorption, and metabolism pathways in CeD pathophysiology and presents potential novel therapeutic opportunities.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

