Over-activation of a nonessential bacterial protease DegP as an antibiotic strategy
- PMID: 33005001
- PMCID: PMC7529758
- DOI: 10.1038/s42003-020-01266-9
Over-activation of a nonessential bacterial protease DegP as an antibiotic strategy
Abstract
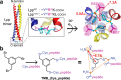
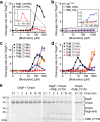
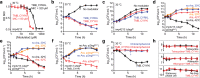
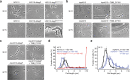
Rising antibiotic resistance urgently begs for novel targets and strategies for antibiotic discovery. Here, we report that over-activation of the periplasmic DegP protease, a member of the highly conserved HtrA family, can be a viable strategy for antibiotic development. We demonstrate that tripodal peptidyl compounds that mimic DegP-activating lipoprotein variants allosterically activate DegP and inhibit the growth of an Escherichia coli strain with a permeable outer membrane in a DegP-dependent fashion. Interestingly, these compounds inhibit bacterial growth at a temperature at which DegP is not essential for cell viability, mainly by over-proteolysis of newly synthesized proteins. Co-crystal structures show that the peptidyl arms of the compounds bind to the substrate-binding sites of DegP. Overall, our results represent an intriguing example of killing bacteria by activating a non-essential enzyme, and thus expand the scope of antibiotic targets beyond the traditional essential proteins or pathways.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

