FUBP1 and FUBP2 enforce distinct epigenetic setpoints for MYC expression in primary single murine cells
- PMID: 33005010
- PMCID: PMC7530719
- DOI: 10.1038/s42003-020-01264-x
FUBP1 and FUBP2 enforce distinct epigenetic setpoints for MYC expression in primary single murine cells
Abstract
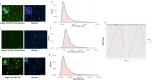
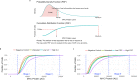
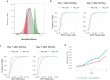
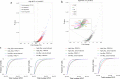
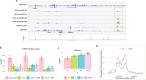
Physiologically, MYC levels must be precisely set to faithfully amplify the transcriptome, but in cancer MYC is quantitatively misregulated. Here, we study the variation of MYC amongst single primary cells (B-cells and murine embryonic fibroblasts, MEFs) for the repercussions of variable cellular MYC-levels and setpoints. Because FUBPs have been proposed to be molecular "cruise controls" that constrain MYC expression, their role in determining basal or activated MYC-levels was also examined. Growing cells remember low and high-MYC setpoints through multiple cell divisions and are limited by the same expression ceiling even after modest MYC-activation. High MYC MEFs are enriched for mRNAs regulating inflammation and immunity. After strong stimulation, many cells break through the ceiling and intensify MYC expression. Lacking FUBPs, unstimulated MEFs express levels otherwise attained only with stimulation and sponsor MYC chromatin changes, revealed by chromatin marks. Thus, the FUBPs enforce epigenetic setpoints that restrict MYC expression.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

