Exosomal Circ-MEMO1 Promotes the Progression and Aerobic Glycolysis of Non-small Cell Lung Cancer Through Targeting MiR-101-3p/KRAS Axis
- PMID: 33005174
- PMCID: PMC7483554
- DOI: 10.3389/fgene.2020.00962
Exosomal Circ-MEMO1 Promotes the Progression and Aerobic Glycolysis of Non-small Cell Lung Cancer Through Targeting MiR-101-3p/KRAS Axis
Abstract
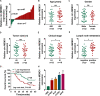
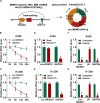
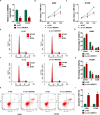
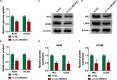
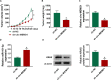
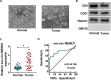
Circular RNA mediator of cell motility 1 (circ-MEMO1) was identified as an oncogene in non-small cell lung cancer (NSCLC). Nevertheless, the working mechanism behind circ-MEMO1-mediated progression of NSCLC is barely known. Quantitative real-time polymerase chain reaction (qRT-PCR) was applied to detect the expression of circ-MEMO1, microRNA-101-3p (miR-101-3p), and KRAS proto-oncogene, GTPase (KRAS). Cell proliferation and aerobic glycolysis were detected by 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay and glycolysis detection kits. Flow cytometry was used to evaluate cell cycle progression and apoptosis of NSCLC cells. Western blot assay was used to measure the protein expression of hexokinase 2 (HK2), lactate dehydrogenase A (LDHA), KRAS, CD9, CD81, tumor susceptibility 101 (TSG101), and Golgi matrix protein 130 kDa (GM130). The target relationship between miR-101-3p and circ-MEMO1 or KRAS was predicted by StarBase software and confirmed by dual-luciferase reporter assay, RNA immunoprecipitation (RIP) assay, and RNA-pull down assay. In vivo tumor growth assay was conducted to assess the effect of circ-MEMO1 in vivo. Exosomes were isolated using the ExoQuick precipitation kit. Circ-MEMO1 was up-regulated in NSCLC, and high expression of circ-MEMO1 predicted poor prognosis in NSCLC patients. Circ-MEMO1 accelerated the proliferation, cell cycle progression, and glycolytic metabolism and inhibited the apoptosis of NSCLC cells. Circ-MEMO1 negatively regulated the expression of miR-101-3p through direct interaction, and si-circ-MEMO1-induced biological effects were attenuated by the introduction of anti-miR-101-3p. MiR-101-3p directly interacted with the 3' untranslated region (3' UTR) of KRAS messenger RNA (mRNA), and KRAS level was regulated by circ-MEMO1/miR-101-3p axis. Circ-MEMO1 silencing suppressed the NSCLC tumor growth in vivo. ROC curve analysis revealed that high expression of serum exosomal circ-MEMO1 (exo-circ-MEMO1) might be a valuable diagnostic marker for NSCLC. Circ-MEMO1 facilitated the progression and glycolysis of NSCLC through regulating miR-101-3p/KRAS axis.
Keywords: KRAS; NSCLC; circ-MEMO1; exosome; glycolysis; miR-101-3p.
Copyright © 2020 Ding, Xi, Wang, Cui, Zhang, Wang, Jiang, Song, Xu and Wang.
Figures



References
-
- Cui Y., Zhang F., Zhu C., Geng L., Tian T., Liu H. (2017). Upregulated lncRNA SNHG1 contributes to progression of non-small cell lung cancer through inhibition of miR-101-3p and activation of Wnt/beta-catenin signaling pathway. Oncotarget 8 17785–17794. 10.18632/oncotarget.14854 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

