Development of an Immune-Related Risk Signature for Predicting Prognosis in Lung Squamous Cell Carcinoma
- PMID: 33005178
- PMCID: PMC7485220
- DOI: 10.3389/fgene.2020.00978
Development of an Immune-Related Risk Signature for Predicting Prognosis in Lung Squamous Cell Carcinoma
Abstract
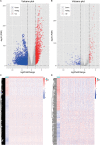
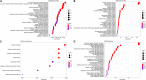
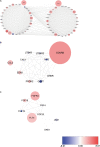
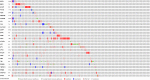
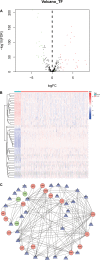
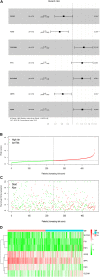
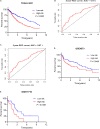
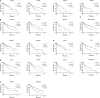
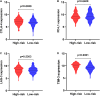
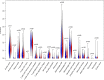
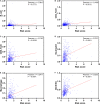
Lung squamous cell carcinoma (LSCC) is the most common subtype of non-small cell lung cancer. Immunotherapy has become an effective treatment in recent years, while patients showed different responses to the current treatment. It is vital to identify the potential immunogenomic signatures to predict patient' prognosis. The expression profiles of LSCC patients with the clinical information were downloaded from TCGA database. Differentially expressed immune-related genes (IRGs) were extracted using edgeR algorithm, and functional enrichment analysis showed that these IRGs were primarily enriched in inflammatory- and immune-related processes. "Cytokine-cytokine receptor interaction" and "PI3K-AKT signaling pathway" were the most enriched KEGG pathways. 27 differentially expressed IRGs were significantly correlated with the overall survival (OS) of patients using univariate Cox regression analysis. A prognostic risk signature that comprises seven IRGs (GCCR, FGF8, CLEC4M, PTH, SLC10A2, NPPC, and FGF4) was developed with effective predictive performance by multivariable Cox stepwise regression analysis. Most importantly, the signature could be an independent prognostic predictor after adjusting for clinicopathological parameters, and also validated in two independent LSCC cohorts (GSE4573 and GSE17710). Potential molecular mechanisms and tumor immune landscape of these IRGs were investigated through computational biology. Analysis of tumor infiltrating lymphocytes and immune checkpoint molecules revealed distinct immune landscape in high- and low-risk group. The study was the first time to construct IRG-based immune signature in the recognition of disease progression and prognosis of LSCC patients.
Keywords: immune-related genes; lung squamous cell carcinoma; prognosis; risk score; signature.
Copyright © 2020 Fu, Zhang, Yang, Huang and Xin.
Figures

References
-
- Ahern E., Cubitt A., Ballard E., Teng M. W. L., Dougall W. C., Smyth M. J., et al. (2019). Pharmacodynamics of Pre-Operative PD1 checkpoint blockade and receptor activator of NFkB ligand (RANKL) inhibition in non-small cell lung cancer (NSCLC): study protocol for a multicentre, open-label, phase 1B/2, translational trial (POPCORN). Trials 20:753. - PMC - PubMed
-
- Brabcova E., Kolesar L., Thorburn E., Striz I. (2014). Chemokines induced in human respiratory epithelial cells by IL-1 family of cytokines. Folia Biol. 60 180–186. - PubMed
LinkOut - more resources
Full Text Sources

