Identification of an Immune-Related Prognostic Signature Associated With Immune Infiltration in Melanoma
- PMID: 33005180
- PMCID: PMC7484056
- DOI: 10.3389/fgene.2020.01002
Identification of an Immune-Related Prognostic Signature Associated With Immune Infiltration in Melanoma
Abstract
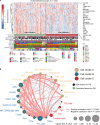
Melanoma is the leading cause of cancer-related death among skin tumors, with an increasing incidence worldwide. Few studies have effectively investigated the significance of an immune-related gene (IRG) signature for melanoma prognosis. Here, we constructed an IRGs prognostic signature using bioinformatics methods and evaluated and validated its predictive capability. Then, immune cell infiltration and tumor mutation burden (TMB) landscapes associated with this signature in melanoma were analyzed comprehensively. With the 10-IRG prognostic signature, melanoma patients in the low-risk group showed better survival with distinct features of high immune cell infiltration and TMB. Importantly, melanoma patients in this subgroup were significantly responsive to MAGE-A3 in the validation cohort. This immune-related prognostic signature is thus a reliable tool to predict melanoma prognosis; as the underlying mechanism of this signature is associated with immune infiltration and mutation burden, it might reflect the benefit of immunotherapy to patients.
Keywords: IRGs; TMB; immune cells infiltration; immunotherapy; melanoma; prognostic signature.
Copyright © 2020 Liu, Liu, Liu, Duan, Huang, Jin, Niu, Zhang and Chen.
Figures






References
LinkOut - more resources
Full Text Sources

