Gene flow as a simple cause for an excess of high-frequency-derived alleles
- PMID: 33005222
- PMCID: PMC7513730
- DOI: 10.1111/eva.12998
Gene flow as a simple cause for an excess of high-frequency-derived alleles
Abstract
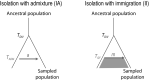
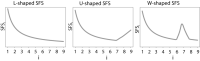
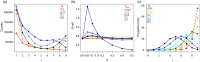
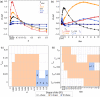
Most human populations exhibit an excess of high-frequency variants, leading to a U-shaped site-frequency spectrum (uSFS). This pattern has been generally interpreted as a signature of ongoing episodes of positive selection, or as evidence for a mis-assignment of ancestral/derived allelic states, but uSFS has also been observed in populations receiving gene flow from a ghost population, in structured populations, or after range expansions. In order to better explain the prevalence of high-frequency variants in humans and other populations, we describe here which patterns of gene flow and population demography can lead to uSFS by using extensive coalescent simulations. We find that uSFS can often be observed in a population if gene flow brings a few ancestral alleles from a well-differentiated population. Gene flow can either consist in single pulses of admixture or continuous immigration, but different demographic conditions are necessary to observe uSFS in these two scenarios. Indeed, an extremely low and recent gene flow is required in the case of single admixture events, while with continuous immigration, uSFS occurs only if gene flow started recently at a high rate or if it lasted for a long time at a low rate. Overall, we find that a neutral uSFS occurs under more restrictive conditions in populations having received single pulses of gene flow than in populations exposed to continuous gene flow. We also show that the uSFS observed in human populations from the 1000 Genomes Project can easily be explained by gene flow from surrounding populations without requiring past episodes of positive selection. These results imply that uSFS should be common in non-isolated populations, such as most wild or domesticated plants and animals.
Keywords: computer simulation; demographic analysis; gene flow; human genetics; human genome; natural selection; neutral evolution.
© 2020 The Authors. Evolutionary Applications published by John Wiley & Sons Ltd.
Conflict of interest statement
None declared.
Figures
References
LinkOut - more resources
Full Text Sources
Miscellaneous

