Global transcriptome study of Dip2B-deficient mouse embryonic lung fibroblast reveals its important roles in cell proliferation and development
- PMID: 33005301
- PMCID: PMC7502710
- DOI: 10.1016/j.csbj.2020.08.030
Global transcriptome study of Dip2B-deficient mouse embryonic lung fibroblast reveals its important roles in cell proliferation and development
Abstract
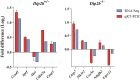
Disco-interacting protein 2 homolog B (Dip2B) is a member of Dip2 family encoded by Dip2b gene. Dip2B has been reported to regulate murine epithelial KIT+ progenitor cell expansion and differentiation epigenetically via exosomal miRNA targeting during salivary gland organogenesis. However, its molecular functions, cellular activities and biological process remain unstudied. Here, we investigated the transcriptome of Dip2B-deficient mouse embryonic lung fibroblasts (MELFs) isolated from E14.5 embryos by RNA-Seq. Expression profiling identified 1369 and 1104 differentially expressed genes (DEGs) from Dip2b-/- and Dip2b+/- MELFs in comparisons to wild-type (Dip2b+/+ ). Functional clustering of DEGs revealed that many gene ontology terms belong to membrane activities such as 'integral component of plasma membrane', and 'ion channel activity', suggesting possible roles of Dip2B in membrane integrity and membrane function. KEGG pathway analysis revealed that multiple metabolic pathways are affected in Dip2b- / - and Dip2b +/ - when compared to Dip2b +/+ MELFs. These include 'protein digestion and absorption', 'pancreatic secretion' and 'steroid hormone synthesis pathway'. These results suggest that Dip2B may play important roles in metabolism. Molecular function analysis shows transcription factors including Hox-genes, bHLH-genes, and Forkhead-genes are significantly down-regulated in Dip2b- / - MELFs. These genes are critical in embryo development and cell differentiation. In addition, Dip2B-deficient MELFs demonstrated a reduction in cell proliferation and migration, and an increase in apoptosis. All results indicate that Dip2B plays multiple roles in cell proliferation, migration and apoptosis during embryogenesis and may participate in control of metabolism. This study provides valuable information for further understanding of the function and regulatory mechanisms of Dip2B.
Keywords: DEG; Dip2B knockout; Gene ontology; KEGG; MELFs; Transcriptome.
© 2020 The Authors.
Figures








References
LinkOut - more resources
Full Text Sources
