3dRNA: Building RNA 3D structure with improved template library
- PMID: 33005304
- PMCID: PMC7508704
- DOI: 10.1016/j.csbj.2020.08.017
3dRNA: Building RNA 3D structure with improved template library
Abstract
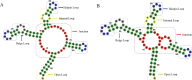
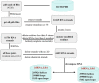
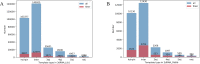
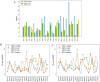
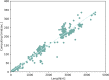
Most of computational methods of building RNA tertiary structure are template-based. The template-based methods usually can give more accurate 3D structures due to the use of native 3D templates, but they cannot work if the 3D templates are not available. So, a more complete library of the native 3D templates is very important for this type of methods. 3dRNA is a template-based method for building RNA tertiary structure previously proposed by us. In this paper we report improved 3D template libraries of 3dRNA by using two different schemes that give two libraries 3dRNA_Lib1 and 3dRNA_Lib2. These libraries expand the original one by nearly ten times. Benchmark shows that they can significantly increase the accuracy of 3dRNA, especially in building complex and large RNA 3D structures.
Keywords: 3D template library; 3dRNA; RNA 3D structure prediction.
© 2020 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


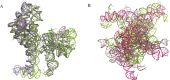
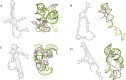
Similar articles
-
3dRNA: 3D Structure Prediction from Linear to Circular RNAs.J Mol Biol. 2022 Jun 15;434(11):167452. doi: 10.1016/j.jmb.2022.167452. Epub 2022 Jan 13. J Mol Biol. 2022. PMID: 35662453
-
3dRNA v2.0: An Updated Web Server for RNA 3D Structure Prediction.Int J Mol Sci. 2019 Aug 23;20(17):4116. doi: 10.3390/ijms20174116. Int J Mol Sci. 2019. PMID: 31450739 Free PMC article.
-
3dRNA/DNA: 3D Structure Prediction from RNA to DNA.J Mol Biol. 2024 Sep 1;436(17):168742. doi: 10.1016/j.jmb.2024.168742. Epub 2024 Aug 7. J Mol Biol. 2024. PMID: 39237199
-
A guide to template based structure prediction.Curr Protein Pept Sci. 2009 Jun;10(3):270-85. doi: 10.2174/138920309788452182. Curr Protein Pept Sci. 2009. PMID: 19519455 Review.
-
The bone regeneration capacity of 3D-printed templates in calvarial defect models: A systematic review and meta-analysis.Acta Biomater. 2019 Jun;91:1-23. doi: 10.1016/j.actbio.2019.04.017. Epub 2019 Apr 11. Acta Biomater. 2019. PMID: 30980937
Cited by
-
Thermodynamic determination of RNA duplex stability in magnesium solutions.Biophys J. 2023 Feb 7;122(3):565-576. doi: 10.1016/j.bpj.2022.12.025. Epub 2022 Dec 20. Biophys J. 2023. PMID: 36540026 Free PMC article.
-
Prediction of 3D RNA Structures from Sequence Using Energy Landscapes of RNA Dimers: Application to RNA Tetraloops.J Chem Theory Comput. 2024 May 28;20(10):4363-4376. doi: 10.1021/acs.jctc.4c00189. Epub 2024 May 10. J Chem Theory Comput. 2024. PMID: 38728627 Free PMC article.
-
RPocket: an intuitive database of RNA pocket topology information with RNA-ligand data resources.BMC Bioinformatics. 2021 Sep 8;22(1):428. doi: 10.1186/s12859-021-04349-4. BMC Bioinformatics. 2021. PMID: 34496744 Free PMC article.
-
Predicting RNA Structure Utilizing Attention from Pretrained Language Models.J Chem Inf Model. 2025 Jul 14;65(13):6483-6498. doi: 10.1021/acs.jcim.4c02094. Epub 2025 Jul 2. J Chem Inf Model. 2025. PMID: 40601389 Free PMC article.
-
Structural Consequences of Deproteinating the 50S Ribosome.Biomolecules. 2022 Oct 31;12(11):1605. doi: 10.3390/biom12111605. Biomolecules. 2022. PMID: 36358955 Free PMC article.
References
-
- Massire C, Westhof E. MANIP: An interactive tool for modelling RNA. J Mol Graph Model 1998;16:197-205.https://doi.org/10.1016/S1093-3263(98)80004-1. - PubMed
LinkOut - more resources
Full Text Sources
