Toward a genetic system in the marine cyanobacterium Prochlorococcus
- PMID: 33005871
- PMCID: PMC7523629
- DOI: 10.1099/acmi.0.000107
Toward a genetic system in the marine cyanobacterium Prochlorococcus
Abstract
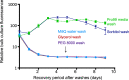
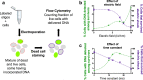
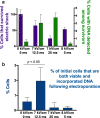
As the smallest and most abundant primary producer in the oceans, the cyanobacterium Prochlorococcus is of interest to diverse branches of science. For the past 30 years, research on this minimal phototroph has led to a growing understanding of biological organization across multiple scales, from the genome to the global ocean ecosystem. Progress in understanding drivers of its diversity and ecology, as well as molecular mechanisms underpinning its streamlined simplicity, has been hampered by the inability to manipulate these cells genetically. Multiple attempts have been made to develop an efficient genetic transformation method for Prochlorococcus over the years; all have been unsuccessful to date, despite some success with their close relative, Synechococcus . To avoid the pursuit of unproductive paths, we report here what has not worked in our hands, as well as our progress developing a method to screen the most efficient electroporation parameters for optimal DNA delivery into Prochlorococcus cells. We also report a novel protocol for obtaining axenic colonies and a new method for differentiating live and dead cells. The electroporation method can be used to optimize DNA delivery into any bacterium, making it a useful tool for advancing transformation systems in other genetically recalcitrant microorganisms.
Keywords: Prochlorococcus; genetic system; intractable bacteria.
© 2020 The Authors.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures

References
-
- Dick GJ, Lam P. Omic approaches to microbial geochemistry. Elements. 2015;11:403–408. doi: 10.2113/gselements.11.6.403. - DOI
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
