Automated digital reporting of clinical laboratory information to national public health surveillance systems, results of a EU/EEA survey, 2018
- PMID: 33006301
- PMCID: PMC7531069
- DOI: 10.2807/1560-7917.ES.2020.25.39.1900591
Automated digital reporting of clinical laboratory information to national public health surveillance systems, results of a EU/EEA survey, 2018
Abstract
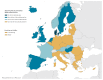
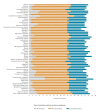
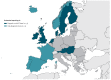
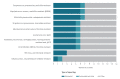
BackgroundTimely reporting of microbiology test results is essential for infection management. Automated, machine-to-machine (M2M) reporting of diagnostic and antimicrobial resistance (AMR) data from laboratory information management systems (LIMS) to public health agencies improves timeliness and completeness of communicable disease surveillance.AimWe surveyed microbiology data reporting practices for national surveillance of EU-notifiable diseases in European Union/European Economic Area (EU/EEA) countries in 2018.MethodsEuropean Centre for Disease Prevention and Control (ECDC) National Microbiology and Surveillance Focal Points completed a questionnaire on the modalities and scope of clinical microbiology laboratory data reporting.ResultsComplete data were provided for all 30 EU/EEA countries. Clinical laboratories used a LIMS in 28 countries. LIMS data on EU-notifiable diseases and AMR were M2M-reported to the national level in 14 and nine countries, respectively. In the 14 countries, associated demographic data reported allowed the de-duplication of patient reports. In 13 countries, M2M-reported data were used for cluster detection at the national level. M2M laboratory data reporting had been validated against conventional surveillance methods in six countries, and replaced those in five. Barriers to M2M reporting included lack of information technology support and financial incentives.ConclusionM2M-reported laboratory data were used for national public health surveillance and alert purposes in nearly half of the EU/EEA countries in 2018. Reported data on infectious diseases and AMR varied in extent and disease coverage across countries and laboratories. Improving automated laboratory-based surveillance will depend on financial and regulatory incentives, and harmonisation of health information and communication systems.
Keywords: antimicrobial resistance; automated data reporting; clinical microbiology testing; communicable diseases; digital surveillance; early-warning; electronic laboratory reporting; public health surveillance.
Conflict of interest statement
Figures
References
-
- Condell O, Gubbels S, Nielsen J, Espenhain L, Frimodt-Møller N, Engberg J, et al. Corrigendum to ‘Automated surveillance system for hospital-acquired urinary tract infections in Denmark’ [Journal of Hospital Infection 93 (2016) 290-296]. J Hosp Infect. 2016;94(4):410. 10.1016/j.jhin.2016.09.001 - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
