4CMenB vaccine induces elite cross-protective human antibodies that compete with human factor H for binding to meningococcal fHbp
- PMID: 33007046
- PMCID: PMC7556464
- DOI: 10.1371/journal.ppat.1008882
4CMenB vaccine induces elite cross-protective human antibodies that compete with human factor H for binding to meningococcal fHbp
Abstract
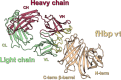
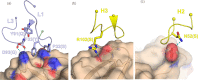
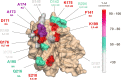
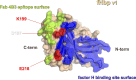
Neisseria meningitidis serogroup B (MenB) is the leading cause of meningococcal meningitis and sepsis in industrialized countries, with the highest incidence in infants and adolescents. Two recombinant protein vaccines that protect against MenB are now available (i.e. 4CMenB and MenB-fHbp). Both vaccines contain the Factor H Binding Protein (fHbp) antigen, which can bind the Human Factor H (fH), the main negative regulator of the alternative complement pathway, thus enabling bacterial survival in the blood. fHbp is present in meningococcal strains as three main variants which are immunologically distinct. Here we sought to obtain detailed information about the epitopes targeted by anti-fHbp antibodies induced by immunization with the 4CMenB multicomponent vaccine. Thirteen anti-fHbp human monoclonal antibodies (mAbs) were identified in a library of over 100 antibody fragments (Fabs) obtained from three healthy adult volunteers immunized with 4CMenB. Herein, the key cross-reactive mAbs were further characterized for antigen binding affinity, complement-mediated serum bactericidal activity (SBA) and the ability to inhibit binding of fH to live bacteria. For the first time, we identified a subset of anti-fHbp mAbs able to elicit human SBA against strains with all three variants and able to compete with human fH for fHbp binding. We present the crystal structure of fHbp v1.1 complexed with human antibody 4B3. The structure, combined with mutagenesis and binding studies, revealed the critical cross-reactive epitope. The structure also provided the molecular basis of competition for fH binding. These data suggest that the fH binding site on fHbp v1.1 can be accessible to the human immune system upon immunization, enabling elicitation of human mAbs broadly protective against MenB. The novel structural, biochemical and functional data are of great significance because the human vaccine-elicited mAbs are the first reported to inhibit the binding of fH to fHbp, and are bactericidal with human complement. Our studies provide molecular insights into the human immune response to the 4CMenB meningococcal vaccine and fuel the rationale for combined structural, immunological and functional studies when seeking deeper understanding of the mechanisms of action of human vaccines.
Conflict of interest statement
I have read the journal's policy and the authors of this manuscript have the following competing interests: All authors except Federica Bianchi were permanent employees of Novartis Vaccines at the time of the study and are now permanent employees of the GSK group of companies. Several authors are listed as inventors on patents owned by the GSK group of companies.
Figures

Similar articles
-
A Meningococcal Outer Membrane Vesicle Vaccine with Overexpressed Mutant FHbp Elicits Higher Protective Antibody Responses in Infant Rhesus Macaques than a Licensed Serogroup B Vaccine.mBio. 2019 Jun 18;10(3):e01231-19. doi: 10.1128/mBio.01231-19. mBio. 2019. PMID: 31213564 Free PMC article.
-
Cocrystal structure of meningococcal factor H binding protein variant 3 reveals a new crossprotective epitope recognized by human mAb 1E6.FASEB J. 2019 Nov;33(11):12099-12111. doi: 10.1096/fj.201900374R. Epub 2019 Oct 5. FASEB J. 2019. PMID: 31442074 Free PMC article.
-
Human factor H (FH) impairs protective meningococcal anti-FHbp antibody responses and the antibodies enhance FH binding.mBio. 2014 Aug 26;5(5):e01625-14. doi: 10.1128/mBio.01625-14. mBio. 2014. PMID: 25161192 Free PMC article.
-
Does binding of complement factor H to the meningococcal vaccine antigen, factor H binding protein, decrease protective serum antibody responses?Clin Vaccine Immunol. 2013 Aug;20(8):1099-107. doi: 10.1128/CVI.00260-13. Epub 2013 Jun 5. Clin Vaccine Immunol. 2013. PMID: 23740919 Free PMC article. Review.
-
Characteristics of a new meningococcal serogroup B vaccine, bivalent rLP2086 (MenB-FHbp; Trumenba®).Postgrad Med. 2016 Aug;128(6):548-56. doi: 10.1080/00325481.2016.1203238. Epub 2016 Jul 7. Postgrad Med. 2016. PMID: 27467048 Review.
Cited by
-
Bactericidal human monoclonal antibody 1B1 shows specificity for meningococcal factor H binding protein variant 2 and displaces human factor H.FASEB Bioadv. 2024 Jun 27;6(8):235-248. doi: 10.1096/fba.2023-00077. eCollection 2024 Aug. FASEB Bioadv. 2024. PMID: 39114449 Free PMC article.
-
Self-assembling protein nanoparticles and virus like particles correctly display β-barrel from meningococcal factor H-binding protein through genetic fusion.PLoS One. 2022 Sep 16;17(9):e0273322. doi: 10.1371/journal.pone.0273322. eCollection 2022. PLoS One. 2022. PMID: 36112575 Free PMC article.
-
Two human antibodies to a meningococcal serogroup B vaccine antigen enhance binding of complement Factor H by stabilizing the Factor H binding site.PLoS Pathog. 2021 Jun 14;17(6):e1009655. doi: 10.1371/journal.ppat.1009655. eCollection 2021 Jun. PLoS Pathog. 2021. PMID: 34125873 Free PMC article.
-
Structural characterization of a cross-protective natural chimera of factor H binding protein from meningococcal serogroup B strain NL096.Comput Struct Biotechnol J. 2022 Apr 18;20:2070-2081. doi: 10.1016/j.csbj.2022.04.011. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35601959 Free PMC article.
-
Design of cross-reactive antigens with machine learning and high-throughput experimental evaluation.Front Bioinform. 2025 Jul 16;5:1580967. doi: 10.3389/fbinf.2025.1580967. eCollection 2025. Front Bioinform. 2025. PMID: 40761757 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

