Array-based analysis of SARS-CoV-2, other coronaviruses, and influenza antibodies in convalescent COVID-19 patients
- PMID: 33007615
- PMCID: PMC7522665
- DOI: 10.1016/j.bios.2020.112643
Array-based analysis of SARS-CoV-2, other coronaviruses, and influenza antibodies in convalescent COVID-19 patients
Abstract
Detection of antibodies to upper respiratory pathogens is critical to surveillance, assessment of the immune status of individuals, vaccine development, and basic biology. The urgent need for antibody detection tools has proven particularly acute in the COVID-19 era. We report a multiplex label-free antigen microarray on the Arrayed Imaging Reflectometry (AIR) platform for detection of antibodies to SARS-CoV-2, SARS-CoV-1, MERS, three circulating coronavirus strains (HKU1, 229E, OC43) and three strains of influenza. We find that the array is readily able to distinguish uninfected from convalescent COVID-19 subjects, and provides quantitative information about total Ig, as well as IgG- and IgM-specific responses.
Keywords: Antigen microarray; Coronavirus; Influenza virus; Label-free biosensor; SARS-CoV-2.
Copyright © 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
B.L.M. is a shareholder of and consultant for Adarza BioSystems, Inc., and is a named inventor on several patents owned by the University of Rochester and licensed to Adarza BioSystems, Inc. D. J. T. is a named inventor on a patent owned by the University of Rochester and licensed to Adarza BioSystems, Inc.
Figures



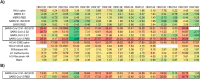

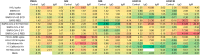

References
-
- Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., Jiang K., Arunkumar G.A., Jurczyszak D., Polanco J., Bermudez-Gonzalez M., Kleiner G., Aydillo T., Miorin L., Fierer D.S., Lugo L.A., Kojic E.M., Stoever J., Liu S.T.H., Cunningham-Rundles C., Feigner P.L., Moran T., Garcia-Sastre A., Caplivski D., Cheng A.C., Kedzierska K., Vapalahti O., Hepojoki J.M., Simon V., Krammer F. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;1–4 - PMC - PubMed
-
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., Marrama D., de Silva A.M., Frazier A., Carlin A., Greenbaum J.A., Peters B., Krammer F., Smith D.M., Crotty S., Sette A. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020 doi: 10.1016/j.cell.2020.05.015. (in press) - DOI - PMC - PubMed
-
- Guo L., Ren L., Yang S., Xiao M., Chang D., Yang F., Dela Cruz C.S., Wang Y., Wu C., Xiao Y., Zhang L., Han L., Dang S., Xu Y., Yang Q.-W., Xu S.-Y., Zhu H.-D., Xu Y.-C., Jin Q., Sharma L., Wang L., Wang J. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa310. (in press) - DOI - PMC - PubMed
-
- Holstein C.A., Griffin M., Hong J., Sampson P.D. Statistical method for determining and comparing limits of detection of bioassays. Anal. Chem. 2015;87:9795–9801. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

