Highly multiplexed quantifications of 299 somatic mutations in colorectal cancer patients by automated MALDI-TOF mass spectrometry
- PMID: 33008377
- PMCID: PMC7532609
- DOI: 10.1186/s12920-020-00804-y
Highly multiplexed quantifications of 299 somatic mutations in colorectal cancer patients by automated MALDI-TOF mass spectrometry
Abstract
Background: Detection of somatic mutations in tumor tissues helps to understand tumor biology and guide treatment selection. Methods such as quantitative PCR can analyze a few mutations with high efficiency, while next generation sequencing (NGS) based methods can analyze hundreds to thousands of mutations. However, there is a lack of cost-effective method for quantitatively analyzing tens to a few hundred mutations of potential biological and clinical significance.
Methods: Through a comprehensive database and literature review we selected 299 mutations associated with colorectal cancer. We then designed a highly multiplexed assay panel (8-wells covering 299 mutations in 109 genes) based on an automated MADLI-TOF mass spectrometry (MS) platform. The multiplex panel was tested with a total of 319 freshly frozen tissues and 92 FFPE samples from 229 colorectal cancer patients, with 13 samples also analyzed by a targeted NGS method covering 532 genes.
Results: Multiplex somatic mutation panel based on MALDI-TOF MS detected and quantified at least one somatic mutation in 142 patients, with KRAS, TP53 and APC being the most frequently mutated genes. Extensive validation by both capillary sequencing and targeted NGS demonstrated high accuracy of the multiplex MS assay. Out of 35 mutations tested with plasmid constructs, sensitivities of 5 and 10% mutant allele frequency were achieved for 19 and 16 mutations, respectively.
Conclusions: Automated MALDI-TOF MS offers an efficient and cost-effective platform for highly multiplexed quantitation of 299 somatic mutations, which may be useful in studying the biological and clinical significance of somatic mutations with large numbers of cancer tissues.
Keywords: Colorectal cancer; MALDI-TOF mass spectrometry; Multiplex detection; Somatic mutation.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

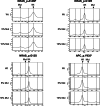
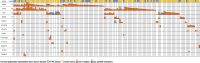
Similar articles
-
Cross-platform comparison of next-generation sequencing and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry for detecting KRAS/NRAS/BRAF/PIK3CA mutations in cfDNA from metastatic colorectal cancer patients.J Clin Lab Anal. 2021 Sep;35(9):e23818. doi: 10.1002/jcla.23818. Epub 2021 Aug 17. J Clin Lab Anal. 2021. PMID: 34403504 Free PMC article.
-
Molecular profiling of lung cancer specimens and liquid biopsies using MALDI-TOF mass spectrometry.Diagn Pathol. 2018 Jan 12;13(1):4. doi: 10.1186/s13000-017-0683-7. Diagn Pathol. 2018. PMID: 29368620 Free PMC article.
-
Panel based MALDI-TOF tumour profiling is a sensitive method for detecting mutations in clinical non small cell lung cancer tumour.PLoS One. 2014 Jun 23;9(6):e100566. doi: 10.1371/journal.pone.0100566. eCollection 2014. PLoS One. 2014. PMID: 24956168 Free PMC article.
-
DNA analysis by MALDI-TOF mass spectrometry.Hum Mutat. 2004 May;23(5):437-41. doi: 10.1002/humu.20023. Hum Mutat. 2004. PMID: 15108274 Review.
-
Application of matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) in the detection of drug resistance of Mycobacterium tuberculosis in re-treated patients.Tuberculosis (Edinb). 2022 Jul;135:102209. doi: 10.1016/j.tube.2022.102209. Epub 2022 Apr 30. Tuberculosis (Edinb). 2022. PMID: 35550524 Review.
Cited by
-
The ideal reporting of RAS testing in colorectal adenocarcinoma: a pathologists' perspective.Pathologica. 2023 Jun 14;115(3):137-47. doi: 10.32074/1591-951X-895. Online ahead of print. Pathologica. 2023. PMID: 37314870 Free PMC article. Review.
-
Molecular mechanism of colorectal cancer and screening of molecular markers based on bioinformatics analysis.Open Life Sci. 2023 Nov 10;18(1):20220687. doi: 10.1515/biol-2022-0687. eCollection 2023. Open Life Sci. 2023. PMID: 37954103 Free PMC article.
References
-
- Soria JC, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, Lee KH, Dechaphunkul A, Imamura F, Nogami N, Kurata T, Okamoto I, Zhou C, Cho BC, Cheng Y, Cho EK, Voon PJ, Planchard D, Su WC, Gray JE, Lee SM, Hodge R, Marotti M, Rukazenkov Y, Ramalingam SS, Investigators F. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung Cancer. N Engl J Med. 2018;378(2):113–125. doi: 10.1056/NEJMoa1713137. - DOI - PubMed
-
- Lievre A, Bachet JB, Boige V, Cayre A, Le Corre D, Buc E, Ychou M, Bouche O, Landi B, Louvet C, Andre T, Bibeau F, Diebold MD, Rougier P, Ducreux M, Tomasic G, Emile JF, Penault-Llorca F, Laurent-Puig P. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol. 2008;26(3):374–379. doi: 10.1200/JCO.2007.12.5906. - DOI - PubMed
-
- Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, Patterson SD, Chang DD. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26(10):1626–1634. doi: 10.1200/JCO.2007.14.7116. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- No. 81672922/National Natural Sciences Foundation of China/International
- No. 2017174160/Medical Health Science and Technology Project of Zhejiang Provincial Health Commission/International
- No. Y20160044/Wenzhou Municipal Science and Technology Bureau/International
- No. 437201702G/Innovation Discipline of Zhejiang Province in Nucleic Acid Molecular Diagnostics/International
- No.437601607/Key Discipline of Zhejiang Province in Medical Technology (First Class, Category A)/International
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

