Viral ecogenomics across the Porifera
- PMID: 33008461
- PMCID: PMC7532657
- DOI: 10.1186/s40168-020-00919-5
Viral ecogenomics across the Porifera
Abstract
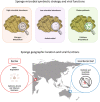
Background: Viruses directly affect the most important biological processes in the ocean via their regulation of prokaryotic and eukaryotic populations. Marine sponges form stable symbiotic partnerships with a wide diversity of microorganisms and this high symbiont complexity makes them an ideal model for studying viral ecology. Here, we used morphological and molecular approaches to illuminate the diversity and function of viruses inhabiting nine sponge species from the Great Barrier Reef and seven from the Red Sea.
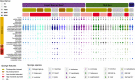
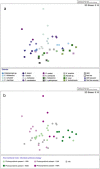
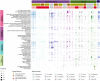
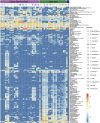
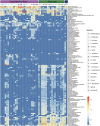
Results: Viromic sequencing revealed host-specific and site-specific patterns in the viral assemblages, with all sponge species dominated by the bacteriophage order Caudovirales but also containing variable representation from the nucleocytoplasmic large DNA virus families Mimiviridae, Marseilleviridae, Phycodnaviridae, Ascoviridae, Iridoviridae, Asfarviridae and Poxviridae. Whilst core viral functions related to replication, infection and structure were largely consistent across the sponge viromes, functional profiles varied significantly between species and sites largely due to differential representation of putative auxiliary metabolic genes (AMGs) and accessory genes, including those associated with herbicide resistance, heavy metal resistance and nylon degradation. Furthermore, putative AMGs varied with the composition and abundance of the sponge-associated microbiome. For instance, genes associated with antimicrobial activity were enriched in low microbial abundance sponges, genes associated with nitrogen metabolism were enriched in high microbial abundance sponges and genes related to cellulose biosynthesis were enriched in species that host photosynthetic symbionts.
Conclusions: Our results highlight the diverse functional roles that viruses can play in marine sponges and are consistent with our current understanding of sponge ecology. Differential representation of putative viral AMGs and accessory genes across sponge species illustrate the diverse suite of beneficial roles viruses can play in the functional ecology of these complex reef holobionts. Video Abstract.
Keywords: AMGs; Coral reef sponges; Functional diversity; Viral ecology; Viromics.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
-
- Bell JJ. The functional roles of marine sponges. Estuar Coast Shelf Sci. 2008;79:341–353. doi: 10.1016/j.ecss.2008.05.002. - DOI
-
- Thomassen S, Riisgård H. Growth and energetics of the sponge Halichondria panicea. Mar Ecol Prog Ser. 1995;128:239–246. doi: 10.3354/meps128239. - DOI
-
- Patterson MR, Chernykh VI, Fialkov VA, Savarese M. Trophic effects of sponge feeding within Lake Baikal’s littoral zone. 1. Insitu pumping rates. Limnol Oceanogr. 1997;42:171–178. doi: 10.4319/lo.1997.42.1.0171. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

