Keratin 17 regulates nuclear morphology and chromatin organization
- PMID: 33008845
- PMCID: PMC7648610
- DOI: 10.1242/jcs.254094
Keratin 17 regulates nuclear morphology and chromatin organization
Abstract
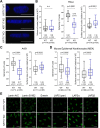
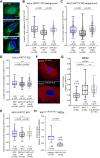
Keratin 17 (KRT17; K17), a non-lamin intermediate filament protein, was recently found to occur in the nucleus. We report here on K17-dependent differences in nuclear morphology, chromatin organization, and cell proliferation. Human tumor keratinocyte cell lines lacking K17 exhibit flatter nuclei relative to normal. Re-expression of wild-type K17, but not a mutant form lacking an intact nuclear localization signal (NLS), rescues nuclear morphology in KRT17-null cells. Analyses of primary cultures of skin keratinocytes from a mouse strain expressing K17 with a mutated NLS corroborated these findings. Proteomics screens identified K17-interacting nuclear proteins with known roles in gene expression, chromatin organization and RNA processing. Key histone modifications and LAP2β (an isoform encoded by TMPO) localization within the nucleus are altered in the absence of K17, correlating with decreased cell proliferation and suppression of GLI1 target genes. Nuclear K17 thus impacts nuclear morphology with an associated impact on chromatin organization, gene expression, and proliferation in epithelial cells.This article has an associated First Person interview with the first author of the paper.
Keywords: Chromatin; Gene expression; Intermediate filament; Keratin; Nucleus; Proliferation.
© 2020. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

