NuA3 HAT antagonizes the Rpd3S and Rpd3L HDACs to optimize mRNA and lncRNA expression dynamics
- PMID: 33010166
- PMCID: PMC7641726
- DOI: 10.1093/nar/gkaa781
NuA3 HAT antagonizes the Rpd3S and Rpd3L HDACs to optimize mRNA and lncRNA expression dynamics
Abstract
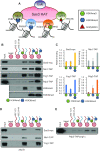
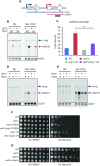
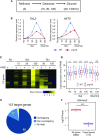
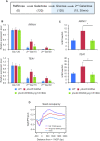
In yeast, NuA3 histone acetyltransferase (NuA3 HAT) promotes acetylation of histone H3 lysine 14 (H3K14) and transcription of a subset of genes through interaction between the Yng1 plant homeodomain (PHD) finger and H3K4me3. Although NuA3 HAT has multiple chromatin binding modules with distinct specificities, their interdependence and combinatorial actions in chromatin binding and transcription remain unknown. Modified peptide pulldown assays reveal that the Yng1 N-terminal region is important for the integrity of NuA3 HAT by mediating the interaction between core subunits and two methyl-binding proteins, Yng1 and Pdp3. We further uncover that NuA3 HAT contributes to the regulation of mRNA and lncRNA expression dynamics by antagonizing the histone deacetylases (HDACs) Rpd3S and Rpd3L. The Yng1 N-terminal region, the Nto1 PHD finger and Pdp3 are important for optimal induction of mRNA and lncRNA transcription repressed by the Set2-Rpd3S HDAC pathway, whereas the Yng1 PHD finger-H3K4me3 interaction affects transcriptional repression memory regulated by Rpd3L HDAC. These findings suggest that NuA3 HAT uses distinct chromatin readers to compete with two Rpd3-containing HDACs to optimize mRNA and lncRNA expression dynamics.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Kouzarides T. Chromatin modifications and their function. Cell. 2007; 128:693–705. - PubMed
-
- Li B., Carey M., Workman J.L.. The role of chromatin during transcription. Cell. 2007; 128:707–719. - PubMed
-
- Shi X., Kachirskaia I., Walter K.L., Kuo J.H., Lake A., Davrazou F., Chan S.M., Martin D.G., Fingerman I.M., Briggs S.D. et al. .. Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J. Biol. Chem. 2007; 282:2450–2455. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

