Molecular and serological characterization of SARS-CoV-2 infection among COVID-19 patients
- PMID: 33011520
- PMCID: PMC7521453
- DOI: 10.1016/j.virol.2020.09.008
Molecular and serological characterization of SARS-CoV-2 infection among COVID-19 patients
Abstract
Background: SARS-CoV-2 is a novel coronavirus and the cause of COVID-19. More than 80% of COVID-19 patients exhibit mild or moderate symptoms. In this study, we investigated the dynamics of viral load and antibodies against SARS-CoV-2 in a longitudinal cohort of COVID-19 patients with severe and mild/moderate diseases.
Methods: Demographic and clinical information were obtained. Serial samples of blood, nasal and pharyngeal and anal swabs were collected at different time points post-onset. SARS-CoV-2 RNA and anti-SARS-CoV-2 antibodies were measured by qRT-PCR and immunoassays, respectively.
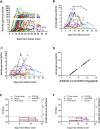
Results: Respiratory SARS-CoV-2 RNA was detectable in 58.0% (58/100) COVID-19 patients upon admission and lasted for a median of 13 days post-onset. In addition, 5.9% (1/17) and 20.2% (19/94) of the blood and anal swab specimens were positive for SARS-CoV-2 RNA, respectively. Anal viral RNA was more frequently detected in the patients who were positive for viral RNA in the respiratory samples upon admission. Specific anti-SARS-CoV-2 antibody developed within two weeks after onset, reached peak approximately 17 days post-onset and then maintained at relatively high level up to 50 days we analyzed in most patients. However, the levels of antibodies were variable among the patients. High titers of antibodies appeared to be associated with the severity of the disease. Furthermore, viral proteins from different sources showed significant difference of serological sensitivity especially during the first week post-onset.
Conclusions: Our results indicate rapid clearance or self-elimination of viral RNA in about half of the COVID-19 patients upon admission. Viral RNA shedding of SARS-CoV-2 occurred in multiple tissues including the respiratory system, blood, and intestine. Variable levels of specific anti-SARS-CoV-2 antibody may be associated with disease severity. These findings have shed light on viral kinetics and antibody response in COVID-19 patients and provide scientific evidence for infection control and patient management.
Keywords: Anti-SARS-CoV-2 antibodies; COVID-19; SARS-CoV-2; Viral RNA.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
We declare no competing interests.
Figures


References
-
- Chan J.F., Yuan S., Kok K.H., To K.K., Chu H., Yang J., Xing F., Liu J., Yip C.C., Poon R.W., Tsoi H.W., Lo S.K., Chan K.H., Poon V.K., Chan W.M., Ip J.D., Cai J.P., Cheng V.C., Chen H., Hui C.K., Yuen K.Y. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. - PMC - PubMed
-
- Chen C., Gao G., Xu Y., Pu L., Wang Q., Wang L., Wang W., Song Y., Chen M., Wang L., Yu F., Yang S., Tang Y., Zhao L., Wang H., Wang Y., Zeng H., Zhang F. SARS-CoV-2-Positive sputum and feces after conversion of pharyngeal samples in patients with COVID-19. Ann. Intern. Med. 2020;172:832–834. - PMC - PubMed
-
- Chen W., Lan Y., Yuan X., Deng X., Li Y., Cai X., Li L., He R., Tan Y., Deng X., Gao M., Tang G., Zhao L., Wang J., Fan Q., Wen C., Tong Y., Tang Y., Hu F., Li F., Tang X. Detectable 2019-nCoV viral RNA in blood is a strong indicator for the further clinical severity. Emerg. Microb. Infect. 2020;9:469–473. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

