Cancer diagnostics based on plasma protein biomarkers: hard times but great expectations
- PMID: 33012111
- PMCID: PMC8169444
- DOI: 10.1002/1878-0261.12809
Cancer diagnostics based on plasma protein biomarkers: hard times but great expectations
Abstract
Cancer diagnostics based on the detection of protein biomarkers in blood has promising potential for early detection and continuous monitoring of disease. However, the currently available protein biomarkers and assay formats largely fail to live up to expectations, mainly due to insufficient diagnostic specificity. Here, we discuss what kinds of plasma proteins might prove useful as biomarkers of malignant processes in specific organs. We consider the need to search for biomarkers deep down in the lowest reaches of the proteome, below current detection levels. In this regard, we comment on the poor molecular detection sensitivity of current protein assays compared to nucleic acid detection reactions, and we discuss requirements for achieving detection of vanishingly small amounts of proteins, to ensure detection of early stages of malignant growth through liquid biopsy.
Keywords: affinity-based protein assays; blood markers; liquid biopsy; protein biomarkers; proximity ligation assay; tissue-specific expression.
© 2020 The Authors. Molecular Oncology published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
Ulf Landegren is a co‐founder and shareholder of Olink Proteomics and Navinci Diagnostics, having rights to the proximity extension assay (PEA) and proximity ligation assay (PLA) technologies.
Figures


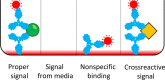
References
-
- Taira A, Merrick G, Wallner K & Dattoli M (2007) Reviving the acid phosphatase test for prostate cancer. Oncology (Williston Park) 21, 1003–1010. - PubMed
-
- Duffy MJ, McDermott EW & Crown J (2018) Blood‐based biomarkers in breast cancer: from proteins to circulating tumor cells to circulating tumor DNA. Tumour Biol 40, 1010428318776169. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

