Metagenomics of the modern and historical human oral microbiome with phylogenetic studies on Streptococcus mutans and Streptococcus sobrinus
- PMID: 33012228
- PMCID: PMC7702799
- DOI: 10.1098/rstb.2019.0573
Metagenomics of the modern and historical human oral microbiome with phylogenetic studies on Streptococcus mutans and Streptococcus sobrinus
Abstract
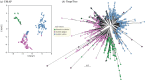
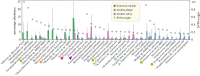
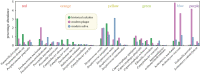
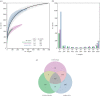
We have recently developed bioinformatic tools to accurately assign metagenomic sequence reads to microbial taxa: SPARSE for probabilistic, taxonomic classification of sequence reads; EToKi for assembling and polishing genomes from short-read sequences; and GrapeTree, a graphic visualizer of genetic distances between large numbers of genomes. Together, these methods support comparative analyses of genomes from ancient skeletons and modern humans. Here, we illustrate these capabilities with 784 samples from historical dental calculus, modern saliva and modern dental plaque. The analyses revealed 1591 microbial species within the oral microbiome. We anticipated that the oral complexes of Socransky et al., which were defined in 1998, would predominate among taxa whose frequencies differed by source. However, although some species discriminated between sources, we could not confirm the existence of the complexes. The results also illustrate further functionality of our pipelines with two species that are associated with dental caries, Streptococcus mutans and Streptococcus sobrinus. They were rare in historical dental calculus but common in modern plaque, and even more common in saliva. Reconstructed draft genomes of these two species from metagenomic samples in which they were abundant were combined with modern public genomes to provide a detailed overview of their core genomic diversity. This article is part of the theme issue 'Insights into health and disease from ancient biomolecules'.
Keywords: ancient DNA; dental calculus; dental plaque; genomic reconstruction; metagenomes; saliva.
Conflict of interest statement
We have no competing interests.
Figures


References
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
