Clinically and biologically relevant subgroups of Wilms tumour defined by genomic and epigenomic analyses
- PMID: 33012783
- PMCID: PMC7853092
- DOI: 10.1038/s41416-020-01102-1
Clinically and biologically relevant subgroups of Wilms tumour defined by genomic and epigenomic analyses
Abstract
Background: Although cure rates for Wilms tumours (WT) are high, many patients receive therapy with attendant long-term complications. Our goal was to stratify WT using genome-wide analyses to identify candidate molecular features for patients who would benefit from a reduction in therapy.
Methods: We generated DNA methylation and exome sequencing data on WT-kidney pairs (n = 57) and unpaired tumours (n = 27) collected either at our centre or by the Children's Oncology Group. Samples were divided into a discovery set (n = 32) and validation set (n = 52).
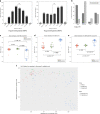
Results: Analysis of DNA methylation revealed two subgroups of WT with distinct features. Subgroup A has a similar DNA methylation profile to mature kidney, while Subgroup B has genome-wide dysregulation of DNA methylation. The rate of non-synonymous missense mutations and segmental chromosomal aberrations was higher in Subgroup B tumours, suggesting that this group has genome instability related to its epigenetic state. Subgroup A had a higher proportion of cases of bilateral disease. Tumours with high-risk histology or from patients who relapsed were only found in Subgroup B.
Conclusion: We have identified subgroup-specific molecular events that could inform future work supporting more targeted therapeutic approaches and patient stratification. We propose a novel developmental tumour model based on these findings.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Chagtai T, Zill C, Dainese L, Wegert J, Savola S, Popov S, et al. Gain of 1q as a prognostic biomarker in Wilms tumors (WTs) treated with preoperative chemotherapy in the International Society of Paediatric Oncology (SIOP) WT 2001 Trial: a SIOP Renal Tumours Biology Consortium Study. J. Clin. Oncol. 2016;34:3195–3203. doi: 10.1200/JCO.2015.66.0001. - DOI - PMC - PubMed
-
- Fernandez CV, Perlman EJ, Mullen EA, Chi Y-Y, Hamilton TE, Gow KW, et al. Clinical outcome and biological predictors of relapse after nephrectomy only for very low-risk Wilms tumor: a report from Children’s Oncology Group AREN0532. Ann. Surg. 2016;265:835–840. doi: 10.1097/SLA.0000000000001716. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

