Comparative mitochondrial genomes of four species of Sinopodisma and phylogenetic implications (Orthoptera, Melanoplinae)
- PMID: 33013166
- PMCID: PMC7515930
- DOI: 10.3897/zookeys.969.49278
Comparative mitochondrial genomes of four species of Sinopodisma and phylogenetic implications (Orthoptera, Melanoplinae)
Abstract
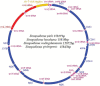
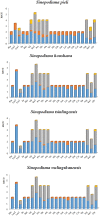
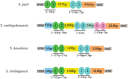
In this study, the whole mitochondrial genomes (mitogenomes) from four species were sequenced. The complete mitochondrial genomes of Sinopodisma pieli, S. houshana, S. qinlingensis, and S. wulingshanensis are 15,857 bp, 15,818 bp, 15,843 bp, and 15,872 bp in size, respectively. The 13 protein-coding genes (PCGs) begin with typical ATN codons, except for COXI in S. qinlingensis, which begins with ACC. The highest A+T content in all the sequenced orthopteran mitogenomes is 76.8% (S. qinlingensis), followed by 76.5% (S. wulingshanensis), 76.4% (S. pieli) and 76.4% (S. houshana) (measured on the major strand). The long polythymine stretches (T-stretch) in the A+T-rich region of the four species are not adjacent to the trnI locus but are inside the stem-loop sequences on the major strand. Moreover, several repeated elements are found in the A+T-rich region of the four species. Phylogenetic analysis based on 53 mitochondrial genomes using Bayesian Inference (BI) and Maximum Likelihood (ML) revealed that Melanoplinae (Podismini) was a monophyletic group; however, the monophyly of Sinopodisma was not supported. These data will provide important information for a better understanding of the phylogenetic relationship of Melanoplinae.
Keywords: Sinopodisma; mitogenome; phylogeny.
Qiu Zhongying, Chang Huihui, Yuan Hao, Huang Yuan, Lu Huimeng, Li Xia, Gou Xingchun.
Figures


References
-
- Cameron SL. (2014) How to sequence and annotate insect mitochondrial genomes for systematic and comparative genomics research. Systematic Entomology 39(3): 400–411. 10.1111/syen.12071 - DOI
-
- Chang H, Qiu Z, Yuan H, Wang X, Li X, Sun H, Guo X, Lu Y, Feng X, Majid M, Huang Y. (2020) Evolutionary rates of and selective constraints on the mitochondrial genomes of Orthoptera insects with different wing types. Molecular Phylogenetics and Evolution 145: 106734. 10.1016/j.ympev.2020.106734 - DOI - PubMed
-
- Chintauan-Marquier IC, Amédégnato C, Nichols RA, Pompanon F, Grandcolas P, Desutter-Grandcolas L. (2014) Inside the Melanoplinae: new molecular evidence for the evolutionary history of the Eurasian Podismini (Orthoptera:Acrididae). Molecular Phylogenetics and Evolution 71: 224–233. 10.1016/j.ympev.2013.09.009 - DOI - PubMed
LinkOut - more resources
Full Text Sources
